Introduction to cloud-optimized bioimaging with OME-Zarr#
Context#
NGFF (Next-generation file formats) is a community driven effort to develop standardized data formats for microscopy that are performant and cloud ready. The primary format is ome.zarr, which is based on the popular zarr n-dimensonial array data format. The development effort is ongoing, but the specification for n-dimensional image data is already present and ready for use. For more information check out the published paper.
Two python packages exist to make life easier for python developers working with the ome.zarr data format:
ome/ome-zarr-py for reading, writing and other functionality
ome/napari-ome-zarr for displaying ome.zarr files in napari.
In this notebook, we will use them to create data in the ome.zarr format, read it back into memory, display it with napari and explore additional functionality.
Background information#
If you are interested to learn more about OME-Zarr, we encourage you to read the associated paper: OME-Zarr: a cloud-optimized bioimaging file format with international community support, Histochemistry and Cell Biology 160(3):1-29, DOI: 10.1007/s00418-023-02209-1, License: CC BY 4.0, July 2023.
Data#
In this episode, we will be using datasets from TBD
Setup#
You can install both packages with pip:
pip install ome-zarr
pip install vizarr
Setup#
This episode uses the following main Python packages:
ome-zarr
vizarr
Please install these packages if not already available in your Python environment.
Packages#
In this episode, Python packages are imported when we start to use them. However, for best software practices, we recommend you to install and import all the necessary libraries at the top of your Jupyter notebook.
pip install ome-zarr vizarr
Show code cell output
Requirement already satisfied: ome-zarr in /srv/conda/envs/notebook/lib/python3.12/site-packages (0.9.0)
Requirement already satisfied: vizarr in /srv/conda/envs/notebook/lib/python3.12/site-packages (0.1.0)
Requirement already satisfied: aiohttp<4 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ome-zarr) (3.10.4)
Requirement already satisfied: dask in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ome-zarr) (2024.8.1)
Requirement already satisfied: distributed in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ome-zarr) (2024.8.1)
Requirement already satisfied: fsspec!=2021.07.0,>=0.8 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from fsspec[s3]!=2021.07.0,>=0.8->ome-zarr) (2024.6.1)
Requirement already satisfied: numpy in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ome-zarr) (1.26.4)
Requirement already satisfied: requests in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ome-zarr) (2.32.3)
Requirement already satisfied: scikit-image in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ome-zarr) (0.24.0)
Requirement already satisfied: toolz in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ome-zarr) (0.12.1)
Requirement already satisfied: zarr>=2.8.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ome-zarr) (2.18.2)
Requirement already satisfied: anywidget in /srv/conda/envs/notebook/lib/python3.12/site-packages (from vizarr) (0.9.13)
Requirement already satisfied: aiohappyeyeballs>=2.3.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from aiohttp<4->ome-zarr) (2.3.6)
Requirement already satisfied: aiosignal>=1.1.2 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from aiohttp<4->ome-zarr) (1.3.1)
Requirement already satisfied: attrs>=17.3.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from aiohttp<4->ome-zarr) (24.2.0)
Requirement already satisfied: frozenlist>=1.1.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from aiohttp<4->ome-zarr) (1.4.1)
Requirement already satisfied: multidict<7.0,>=4.5 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from aiohttp<4->ome-zarr) (6.0.5)
Requirement already satisfied: yarl<2.0,>=1.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from aiohttp<4->ome-zarr) (1.9.4)
Requirement already satisfied: s3fs in /srv/conda/envs/notebook/lib/python3.12/site-packages (from fsspec[s3]!=2021.07.0,>=0.8->ome-zarr) (2024.6.1)
Requirement already satisfied: asciitree in /srv/conda/envs/notebook/lib/python3.12/site-packages (from zarr>=2.8.1->ome-zarr) (0.3.3)
Requirement already satisfied: numcodecs>=0.10.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from zarr>=2.8.1->ome-zarr) (0.12.1)
Requirement already satisfied: fasteners in /srv/conda/envs/notebook/lib/python3.12/site-packages (from zarr>=2.8.1->ome-zarr) (0.17.3)
Requirement already satisfied: ipywidgets>=7.6.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from anywidget->vizarr) (8.1.3)
Requirement already satisfied: psygnal>=0.8.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from anywidget->vizarr) (0.11.1)
Requirement already satisfied: typing-extensions>=4.2.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from anywidget->vizarr) (4.12.2)
Requirement already satisfied: click>=8.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from dask->ome-zarr) (8.1.7)
Requirement already satisfied: cloudpickle>=3.0.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from dask->ome-zarr) (3.0.0)
Requirement already satisfied: packaging>=20.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from dask->ome-zarr) (24.1)
Requirement already satisfied: partd>=1.4.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from dask->ome-zarr) (1.4.2)
Requirement already satisfied: pyyaml>=5.3.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from dask->ome-zarr) (6.0.2)
Requirement already satisfied: jinja2>=2.10.3 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from distributed->ome-zarr) (3.1.4)
Requirement already satisfied: locket>=1.0.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from distributed->ome-zarr) (1.0.0)
Requirement already satisfied: msgpack>=1.0.2 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from distributed->ome-zarr) (1.0.8)
Requirement already satisfied: psutil>=5.8.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from distributed->ome-zarr) (5.9.8)
Requirement already satisfied: sortedcontainers>=2.0.5 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from distributed->ome-zarr) (2.4.0)
Requirement already satisfied: tblib>=1.6.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from distributed->ome-zarr) (3.0.0)
Requirement already satisfied: tornado>=6.2.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from distributed->ome-zarr) (6.4.1)
Requirement already satisfied: urllib3>=1.26.5 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from distributed->ome-zarr) (1.26.19)
Requirement already satisfied: zict>=3.0.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from distributed->ome-zarr) (3.0.0)
Requirement already satisfied: charset-normalizer<4,>=2 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from requests->ome-zarr) (3.3.2)
Requirement already satisfied: idna<4,>=2.5 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from requests->ome-zarr) (3.7)
Requirement already satisfied: certifi>=2017.4.17 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from requests->ome-zarr) (2024.7.4)
Requirement already satisfied: scipy>=1.9 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from scikit-image->ome-zarr) (1.14.0)
Requirement already satisfied: networkx>=2.8 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from scikit-image->ome-zarr) (3.3)
Requirement already satisfied: pillow>=9.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from scikit-image->ome-zarr) (10.4.0)
Requirement already satisfied: imageio>=2.33 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from scikit-image->ome-zarr) (2.34.2)
Requirement already satisfied: tifffile>=2022.8.12 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from scikit-image->ome-zarr) (2024.8.10)
Requirement already satisfied: lazy-loader>=0.4 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from scikit-image->ome-zarr) (0.4)
Requirement already satisfied: comm>=0.1.3 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipywidgets>=7.6.0->anywidget->vizarr) (0.2.2)
Requirement already satisfied: ipython>=6.1.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipywidgets>=7.6.0->anywidget->vizarr) (8.17.2)
Requirement already satisfied: traitlets>=4.3.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipywidgets>=7.6.0->anywidget->vizarr) (5.14.3)
Requirement already satisfied: widgetsnbextension~=4.0.11 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipywidgets>=7.6.0->anywidget->vizarr) (4.0.11)
Requirement already satisfied: jupyterlab-widgets~=3.0.11 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipywidgets>=7.6.0->anywidget->vizarr) (3.0.11)
Requirement already satisfied: MarkupSafe>=2.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from jinja2>=2.10.3->distributed->ome-zarr) (2.1.5)
Requirement already satisfied: aiobotocore<3.0.0,>=2.5.4 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from s3fs->fsspec[s3]!=2021.07.0,>=0.8->ome-zarr) (2.13.2)
Requirement already satisfied: botocore<1.34.132,>=1.34.70 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from aiobotocore<3.0.0,>=2.5.4->s3fs->fsspec[s3]!=2021.07.0,>=0.8->ome-zarr) (1.34.131)
Requirement already satisfied: wrapt<2.0.0,>=1.10.10 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from aiobotocore<3.0.0,>=2.5.4->s3fs->fsspec[s3]!=2021.07.0,>=0.8->ome-zarr) (1.16.0)
Requirement already satisfied: aioitertools<1.0.0,>=0.5.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from aiobotocore<3.0.0,>=2.5.4->s3fs->fsspec[s3]!=2021.07.0,>=0.8->ome-zarr) (0.11.0)
Requirement already satisfied: decorator in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (5.1.1)
Requirement already satisfied: jedi>=0.16 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (0.19.1)
Requirement already satisfied: matplotlib-inline in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (0.1.7)
Requirement already satisfied: prompt-toolkit!=3.0.37,<3.1.0,>=3.0.30 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (3.0.38)
Requirement already satisfied: pygments>=2.4.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (2.18.0)
Requirement already satisfied: stack-data in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (0.6.2)
Requirement already satisfied: pexpect>4.3 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (4.9.0)
Requirement already satisfied: jmespath<2.0.0,>=0.7.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from botocore<1.34.132,>=1.34.70->aiobotocore<3.0.0,>=2.5.4->s3fs->fsspec[s3]!=2021.07.0,>=0.8->ome-zarr) (1.0.1)
Requirement already satisfied: python-dateutil<3.0.0,>=2.1 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from botocore<1.34.132,>=1.34.70->aiobotocore<3.0.0,>=2.5.4->s3fs->fsspec[s3]!=2021.07.0,>=0.8->ome-zarr) (2.8.2)
Requirement already satisfied: parso<0.9.0,>=0.8.3 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from jedi>=0.16->ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (0.8.4)
Requirement already satisfied: ptyprocess>=0.5 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from pexpect>4.3->ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (0.7.0)
Requirement already satisfied: wcwidth in /srv/conda/envs/notebook/lib/python3.12/site-packages (from prompt-toolkit!=3.0.37,<3.1.0,>=3.0.30->ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (0.2.13)
Requirement already satisfied: executing>=1.2.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from stack-data->ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (2.0.1)
Requirement already satisfied: asttokens>=2.1.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from stack-data->ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (2.4.1)
Requirement already satisfied: pure-eval in /srv/conda/envs/notebook/lib/python3.12/site-packages (from stack-data->ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (0.2.3)
Requirement already satisfied: six>=1.12.0 in /srv/conda/envs/notebook/lib/python3.12/site-packages (from asttokens>=2.1.0->stack-data->ipython>=6.1.0->ipywidgets>=7.6.0->anywidget->vizarr) (1.16.0)
Note: you may need to restart the kernel to use updated packages.
Import necessary Python packages#
from ome_zarr.io import parse_url
from ome_zarr.reader import Reader
import zarr
import vizarr
Reading bioimaging OME-Zarr file from Remote source#
When opening and reading the OME-Zarr file, only metadata is effectively loaded into memory.
Data will be loaded whenever needed.
url = "https://uk1s3.embassy.ebi.ac.uk/idr/zarr/v0.4/idr0062A/6001240.zarr"
# read the image data
store = parse_url(url, mode="r").store
reader = Reader(parse_url(url))
# nodes may include images, labels etc
nodes = list(reader())
# first node will be the image pixel data
image_node = nodes[0]
The OME-NGFF/OME-ZARR standard#
Reference: Using Zarr for images – The OME-ZARR standard, blog written by Fabrizio Musacchio in October 24, 2022.
The Open Microscopy Environment (OME) has proposed the next-generation file format (NGFF) specifications (see Specification GitHub repository) for storing multi-resolution bioimaging data in the cloud. The OME defines this OME-NGFF called standard based on the Zarr file format, which provides the necessary support for storing and accessing arrays from distributed cloud storages. The advantages of using such a standard are:
standardization of the metadata (regarding both format and name definition, according the OME-XML structured annotations specifications,
standardization of the structure within the Zarr file (i.e., how the image is stored within the Zarr file, and
standardization of the Zarr-store format (Zarr storage specifications).
This standardization enables developers to easily provide an API, that works for all OME-ZARR files in a unified manner. We can write image analysis pipelines and don’t have to care, how the hierarchy within a Zarr files might be organized – we can expect it to always be arranged and accessible in the same way. The standardization of the metadata also sets a clear frame for how to store and name image attributes such as the microscope metadata, image resolution or the channel specifications.
nodes[0].data
[dask.array<from-zarr, shape=(2, 236, 275, 271), dtype=uint16, chunksize=(1, 1, 275, 271), chunktype=numpy.ndarray>,
dask.array<from-zarr, shape=(2, 236, 137, 135), dtype=uint16, chunksize=(1, 1, 137, 135), chunktype=numpy.ndarray>,
dask.array<from-zarr, shape=(2, 236, 68, 67), dtype=uint16, chunksize=(1, 1, 68, 67), chunktype=numpy.ndarray>]
We see that OME-Zarr consists of three different Dask arrays, each with a different shape, corresponding to the three different resolutions stored to optimize access.
Inspect metadata#
The coordinate transformation can be inspected by printing the metadata information.
image_node.metadata
{'axes': [{'name': 'c', 'type': 'channel'},
{'name': 'z', 'type': 'space', 'unit': 'micrometer'},
{'name': 'y', 'type': 'space', 'unit': 'micrometer'},
{'name': 'x', 'type': 'space', 'unit': 'micrometer'}],
'name': None,
'coordinateTransformations': [[{'scale': [1.0,
0.5002025531914894,
0.3603981534640209,
0.3603981534640209],
'type': 'scale'}],
[{'scale': [1.0, 0.5002025531914894, 0.7207963069280418, 0.7207963069280418],
'type': 'scale'}],
[{'scale': [1.0, 0.5002025531914894, 1.4415926138560835, 1.4415926138560835],
'type': 'scale'}]],
'channel_names': ['LaminB1', 'Dapi'],
'visible': [True, True],
'contrast_limits': [[0.0, 1500.0], [0.0, 1500.0]],
'colormap': [[[0, 0, 0], [0.0, 0.0, 1.0]], [[0, 0, 0], [1.0, 1.0, 0.0]]]}
Visualization with Vizarr#
vizarr allows you to interactively visualize your OME-Zarr and select channels, different Z values, etc. Below is an example of the view you can get:
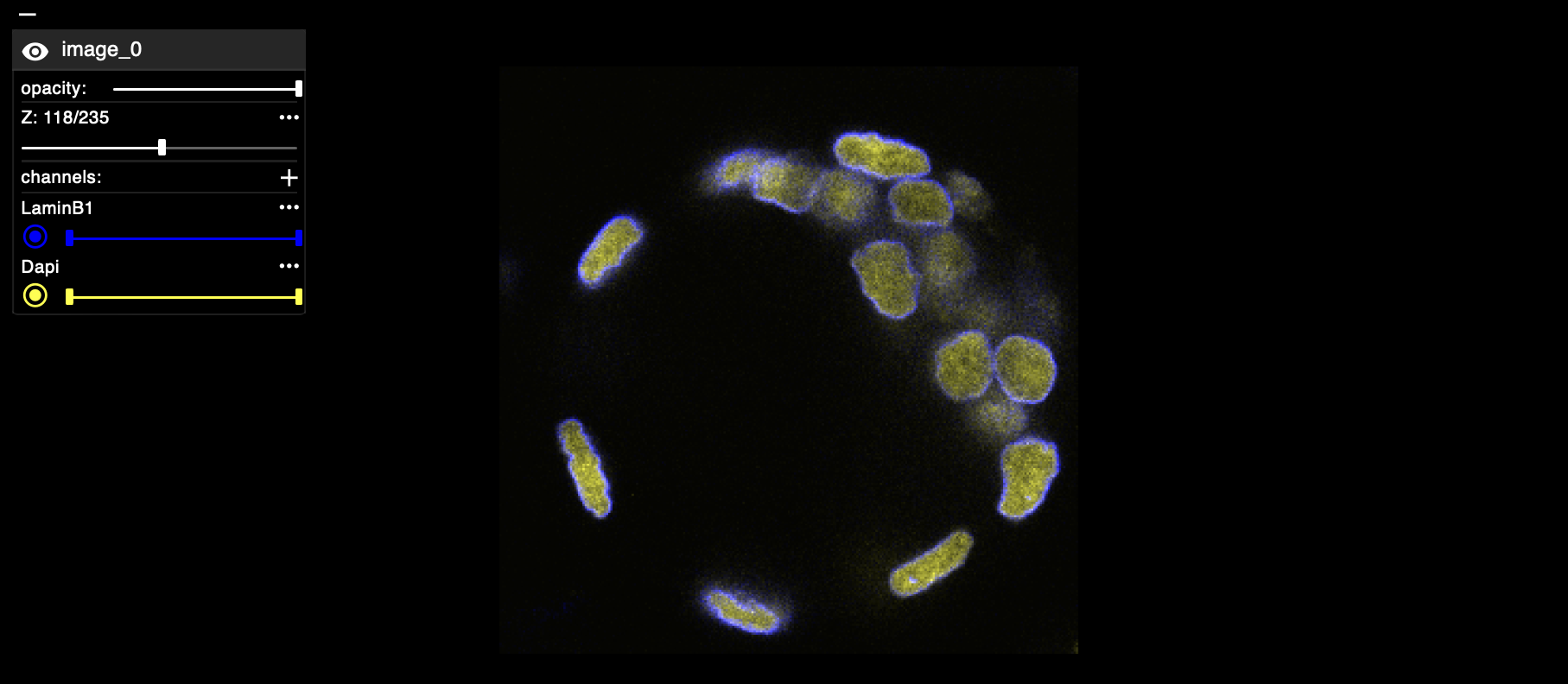
viewer = vizarr.Viewer()
viewer.add_image(store)
viewer
Show code cell output
Using Xarray for Multi-scale OME-Zarr#
OME-Zarr standard is a multiscale zarr which is not handled directly by Xarray yet. Therefore, to use Xarray, you first need to select which “scale/resolution” you wish to access.
from xarray_ome_ngff import read_multiscale_array
array_z = zarr.open_array(url + "/0")
array_x = read_multiscale_array(array_z)
array_x
<xarray.DataArray (c: 2, z: 236, y: 275, x: 271)> Size: 70MB
array([[[[ 8, 9, 8, ..., 9, 9, 10],
[ 9, 9, 9, ..., 8, 9, 9],
[ 8, 8, 8, ..., 26, 40, 8],
...,
[ 9, 9, 9, ..., 9, 10, 14],
[ 8, 9, 10, ..., 9, 10, 9],
[ 9, 8, 10, ..., 10, 8, 8]],
[[ 9, 9, 9, ..., 8, 11, 11],
[ 9, 8, 9, ..., 10, 9, 10],
[ 9, 16, 9, ..., 39, 30, 9],
...,
[10, 9, 10, ..., 10, 10, 9],
[10, 8, 10, ..., 10, 10, 10],
[10, 11, 9, ..., 9, 10, 10]],
[[ 9, 9, 9, ..., 14, 7, 15],
[ 9, 9, 9, ..., 10, 9, 9],
[ 8, 9, 9, ..., 9, 67, 8],
...,
...
...,
[28, 28, 28, ..., 29, 28, 51],
[27, 29, 28, ..., 28, 29, 28],
[28, 37, 28, ..., 28, 28, 52]],
[[28, 29, 27, ..., 31, 30, 37],
[30, 27, 28, ..., 28, 28, 32],
[28, 29, 32, ..., 31, 46, 29],
...,
[29, 27, 29, ..., 28, 36, 29],
[29, 29, 28, ..., 29, 28, 28],
[28, 28, 29, ..., 27, 29, 28]],
[[33, 27, 27, ..., 35, 42, 28],
[28, 28, 28, ..., 29, 28, 43],
[28, 28, 27, ..., 28, 27, 28],
...,
[28, 28, 28, ..., 27, 27, 40],
[28, 27, 29, ..., 28, 31, 27],
[27, 27, 28, ..., 27, 44, 32]]]], dtype=uint16)
Coordinates:
* c (c) float64 16B 0.0 1.0
* z (z) float64 2kB 0.0 0.5002 1.0 1.501 ... 116.0 116.5 117.0 117.5
* y (y) float64 2kB 0.0 0.3604 0.7208 1.081 ... 97.67 98.03 98.39 98.75
* x (x) float64 2kB 0.0 0.3604 0.7208 1.081 ... 96.23 96.59 96.95 97.31Name variable “data” and get Xarray dataset#
dset = array_x.rename("data").to_dataset()
dset
<xarray.Dataset> Size: 70MB
Dimensions: (c: 2, z: 236, y: 275, x: 271)
Coordinates:
* c (c) float64 16B 0.0 1.0
* z (z) float64 2kB 0.0 0.5002 1.0 1.501 ... 116.0 116.5 117.0 117.5
* y (y) float64 2kB 0.0 0.3604 0.7208 1.081 ... 97.67 98.03 98.39 98.75
* x (x) float64 2kB 0.0 0.3604 0.7208 1.081 ... 96.23 96.59 96.95 97.31
Data variables:
data (c, z, y, x) uint16 70MB 8 9 8 10 8 11 9 9 ... 31 29 40 28 27 44 32Assign channel names as coordinates#
channel_names = image_node.metadata["channel_names"]
dset = dset.assign_coords({"c": channel_names})
dset
<xarray.Dataset> Size: 70MB
Dimensions: (z: 236, y: 275, x: 271, c: 2)
Coordinates:
* z (z) float64 2kB 0.0 0.5002 1.0 1.501 ... 116.0 116.5 117.0 117.5
* y (y) float64 2kB 0.0 0.3604 0.7208 1.081 ... 97.67 98.03 98.39 98.75
* x (x) float64 2kB 0.0 0.3604 0.7208 1.081 ... 96.23 96.59 96.95 97.31
* c (c) <U7 56B 'LaminB1' 'Dapi'
Data variables:
data (c, z, y, x) uint16 70MB 8 9 8 10 8 11 9 9 ... 31 29 40 28 27 44 32Select channels and visualize data#
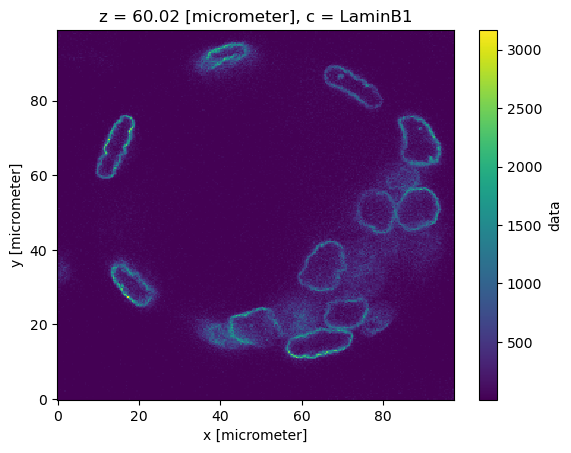
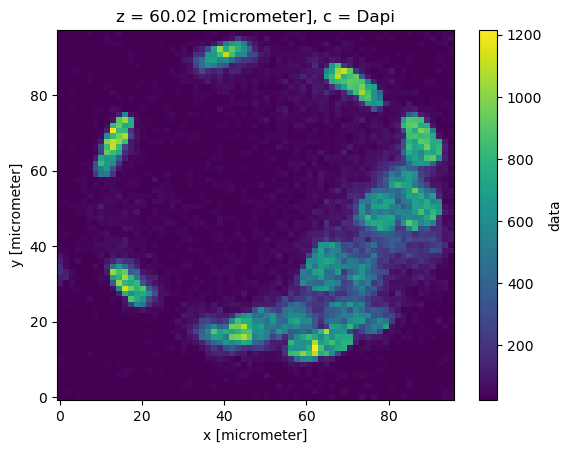
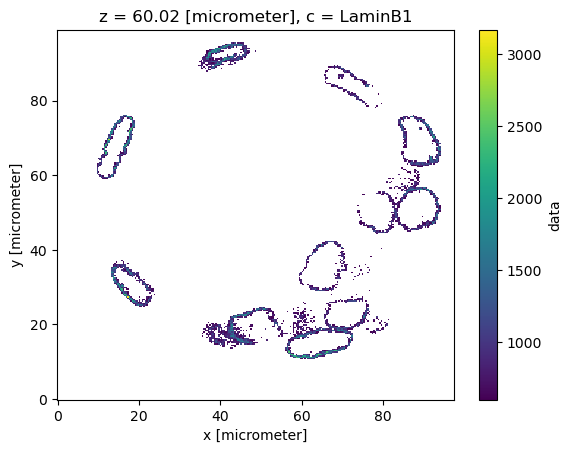
dset.sel(c="LaminB1").isel(z=120)["data"].plot()
<matplotlib.collections.QuadMesh at 0x7f6f109f0410>
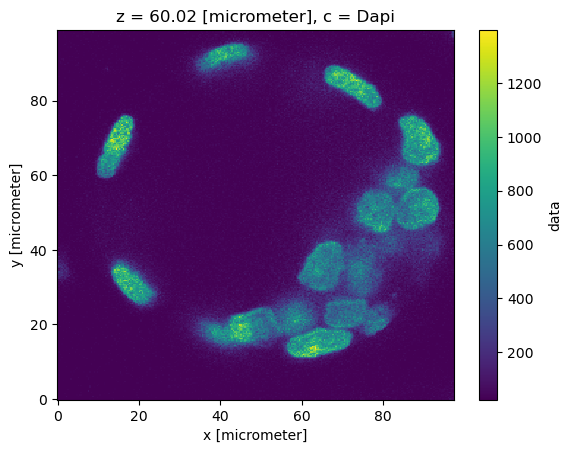
dset.sel(c="Dapi").isel(z=120)["data"].plot()
<matplotlib.collections.QuadMesh at 0x7f6f108b6570>
Reading a different resolution#
read_multiscale_array(zarr.open_array(url + "/1")).rename("data").to_dataset().assign_coords({"c": channel_names}).sel(c="Dapi").isel(z=120)["data"].plot()
<matplotlib.collections.QuadMesh at 0x7f6f02fcbad0>
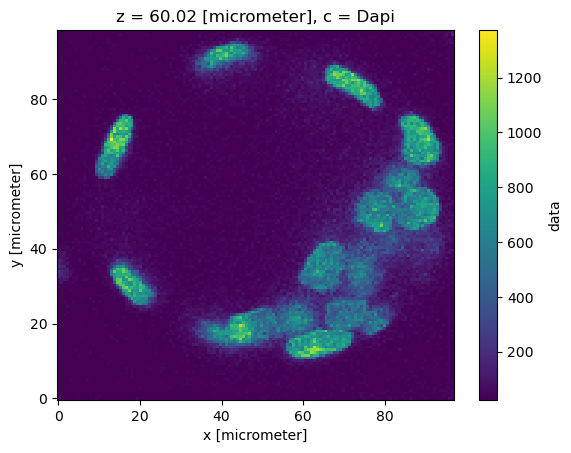
read_multiscale_array(zarr.open_array(url + "/2")).rename("data").to_dataset().assign_coords({"c": channel_names}).sel(c="Dapi").isel(z=120)["data"].plot()
<matplotlib.collections.QuadMesh at 0x7f6f02e63ad0>
Using hvplot to visualize Bio-imaging data#
import holoviews as hv
import hvplot.xarray
import numpy as np
dset.z
<xarray.DataArray 'z' (z: 236)> Size: 2kB
array([ 0. , 0.500203, 1.000405, ..., 116.547195, 117.047397,
117.5476 ])
Coordinates:
* z (z) float64 2kB 0.0 0.5002 1.0 1.501 ... 116.0 116.5 117.0 117.5
Attributes:
units: micrometerdset["data"].sel(z=slice(40,120)).hvplot(x="x", y="y", cmap="RdYlGn", width=600, height=500)
Overlap the two channels on one plot#
dset.where(dset.sel(c="LaminB1") > 600, np.nan).sel(c=["LaminB1"]).isel(z=120)["data"].plot()
<matplotlib.collections.QuadMesh at 0x7f6f00122510>
p1 = dset.sel(c="Dapi").isel(z=120)["data"].hvplot(cmap="RdYlGn", width=600, height=500)
p2 = dset.where(dset.sel(c="LaminB1") > 600, np.nan).sel(c=["LaminB1"]).isel(z=120)["data"].hvplot(x="x", y="y", cmap="binary_r", width=600, height=500)
p1 * p2


