Rapid Landslide Detection using Synthetic Aperture Radar (SAR) Datacubes#
Context#
With the increasing impact of climate change on landslide events, there is a need for rapid landslide detection technologies. Synthetic Aperture Radar (SAR) offers a solution by providing reliable measurements regardless of weather conditions.
The advancement of deep learning (DL) algorithms for SAR applications is still stalled due to its intricate features and the need for extensive preprocessing.
Earth Observation datacubes consist of processed data that can be easily utilized by researchers in remote sensing and machine learning fields. Remote sensing scientists can analyze time-series data of specific geographic areas, while machine learning experts can incorporate the data into their models.
This notebook demonstrate the added value of analysis-ready SAR datasets to train supervised deep learning models for landslide detection.
We demonstrate the use of Dask with xbatcher to parallelize the generation of the training and test partitions.
Modelling approach#
This notebook applies a U-Net architecture for landslide detection using SAR datacubes. The model is trained over an analysis-ready dataset comprising of SAR intensity and interferometry information, accumulated both before and after disaster events that initiated landslides.
The details of the model are given in the paper entitled “Deep Learning for Rapid Landslide Detection using Synthetic Aperture Radar (SAR) Datacubes” [BLM+22].
Data#
We will be using the Hokkaido ARD dataset archived in Zenodo and mirrored for the purpose of this notebook into a remote cloud storage bucket provided by Pangeo-EOSC MinIO. The dataset contains multiple layers of SAR time-series data and interferometric products (coherence, InSAR) and auxiliary topographic information (elevation, slope, aspect, curvature) for multiple landslide events along with the corresponding landslide labels as indicated in the image below.
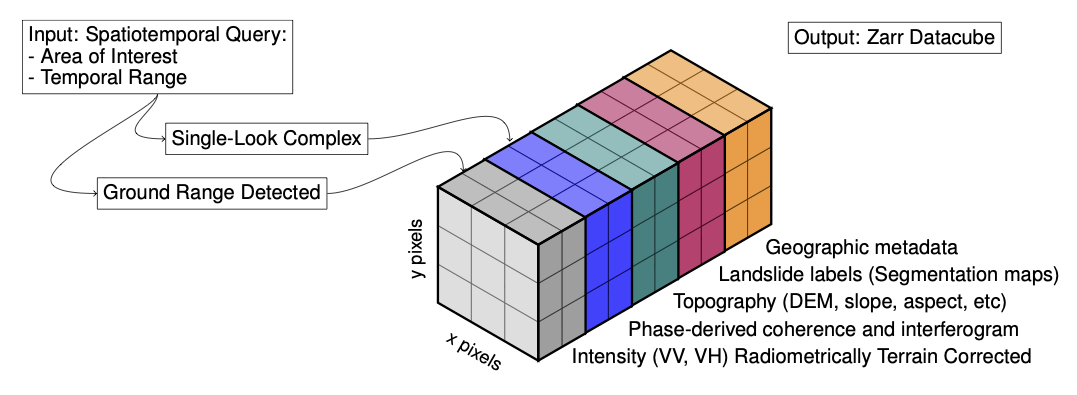
Fig. 1 Datacube containing Synthetic ApertureRadar (SAR) data and other geographic layers. Image credits: [BLM+22].#
In addition to the Hokkaido dataset, there are other analysis-ready datacubes in three geographical areas: Puerto Rico,Mt Talakmau (Indonesia) and Kaikoura (New Zealand).
All datasets are in Zarr format and can be accessed locally through downloading the data from Zenodo or remotely through mirror datasets stored in a MinIO S3 compatible object storage.
Setup#
This episode uses the following main Python packages:
xbatcher [xbatcherDTeam24]
pytorch [PGM+19] with pyTorch lightning [FThePLteam19]
dask [DaskDTeam16]
Please install these packages if not already available in your Python environment.
Packages#
In this episode, some Python packages are imported when we start to use them. However, for best software practices, we recommend you to install and import all the necessary libraries at the top of your Jupyter notebook.
Load libraries#
"""Math & Data Libraries"""
import numpy as np
import xarray as xr
import xbatcher
import s3fs
import pooch
from fsspec.implementations.zip import ZipFileSystem
from fsspec.mapping import FSMap
import io
""" ML Libraries"""
import torch
from torch.utils.data import ConcatDataset, DataLoader, Dataset, random_split
from torchmetrics import AUROC, AveragePrecision, F1Score
import segmentation_models_pytorch as smp
import pytorch_lightning as pl
from pytorch_lightning import LightningDataModule, seed_everything
"""Visualization Libraries"""
import matplotlib.pyplot as plt
""" Miscellaneous Libraries"""
import copy
from typing import Any, Optional, Tuple, List
from tqdm.notebook import tqdm
Create a local Dask cluster on the local machine#
from dask.distributed import Client
client = Client() # create `a local dask cluster on the local machine.
client
Client
Client-88a8955d-3b8b-11ef-8fe8-12e5624d92c1
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:8787/status |
Cluster Info
LocalCluster
7621921f
| Dashboard: http://127.0.0.1:8787/status | Workers: 4 |
| Total threads: 8 | Total memory: 32.00 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-d3b6e427-16f7-454a-9a44-6bd0402d4ee9
| Comm: tcp://127.0.0.1:40621 | Workers: 4 |
| Dashboard: http://127.0.0.1:8787/status | Total threads: 8 |
| Started: Just now | Total memory: 32.00 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:38829 | Total threads: 2 |
| Dashboard: http://127.0.0.1:34181/status | Memory: 8.00 GiB |
| Nanny: tcp://127.0.0.1:46359 | |
| Local directory: /tmp/dask-scratch-space/worker-ycnjjkqz | |
Worker: 1
| Comm: tcp://127.0.0.1:36581 | Total threads: 2 |
| Dashboard: http://127.0.0.1:35555/status | Memory: 8.00 GiB |
| Nanny: tcp://127.0.0.1:36061 | |
| Local directory: /tmp/dask-scratch-space/worker-onwvsv99 | |
Worker: 2
| Comm: tcp://127.0.0.1:33709 | Total threads: 2 |
| Dashboard: http://127.0.0.1:34173/status | Memory: 8.00 GiB |
| Nanny: tcp://127.0.0.1:36617 | |
| Local directory: /tmp/dask-scratch-space/worker-3lh2479w | |
Worker: 3
| Comm: tcp://127.0.0.1:41079 | Total threads: 2 |
| Dashboard: http://127.0.0.1:33039/status | Memory: 8.00 GiB |
| Nanny: tcp://127.0.0.1:43509 | |
| Local directory: /tmp/dask-scratch-space/worker-06rr9pk7 | |
Settings#
Experiments#
The experiments are defined using the hparams dictionary. We’re setting the dataset name (ds_name), the dataset storage (ds_storage), the timestep length (timestep_length), the input variables (input_vars), the patch size (patch_size), the batch size (batch_size), the learning rate (lr), the weight decay (weight_decay), the loss function (loss), and the maximum number of epochs (max_epochs).
In this experiment, we are working with the Hokkaido dataset loaded from a remote storage bucket. We are using SAR bands before and after the earthquake, plus DEM-derived data (elevation, slope, aspect, curvature) are used (total 8 input channels). We will only analyzing a single time step/satellite pass before and after the event. The patch size is set to 128x128 pixels, and the batch size is set to 64. The learning rate is set to 0.01, the weight decay to 0.0001, and the loss function to Cross-Entropy. The maximum number of epochs is set to 30.
Further details of other experiments can be found in the methods paper [BLM+22].
hparams = {
"ds_name":"hokkaido_japan",
"ds_storage":"remote",
"timestep_length":1,
"input_vars": "vv_before,vv_after,vh_before,vh_after,dem,dem_aspect,dem_curvature,dem_slope_radians",
"patch_size": (128, 128),
"batch_size":64,
"lr":0.01,
"weight_decay":0.0001,
"loss":"ce",
"max_epochs":30
}
Data Module#
The functions and classes below, original defined in the modelling codebase, use xarray to access the dataset, xbatcher to facilitate the generation of patches and torch.utils.data modules to define datasets.
In addition to the original code, we have added the open_zarr_source function to access the dataset from the original Zenodo source or the mirrored dataset stored in the MinIO S3 compatible object storage.
# function to access the Zarr dataset
def open_zarr_source(ds_name, ds_storage):
if ds_storage == 'remote':
fst = s3fs.S3FileSystem(anon=True,
client_kwargs={
"endpoint_url": "https://pangeo-eosc-minioapi.vm.fedcloud.eu/"
})
s3_prefix = 'acocacastro2'
s3_suffix = 'igarss24'
s3_bucket = s3_prefix + '-' + s3_suffix + '/'
s3_path = "s3://" + s3_bucket + "/" + ds_name + ".zarr.zip"
f = fst.open(s3_path)
fs = ZipFileSystem(f, mode="r")
store = FSMap("", fs, check=False)
ds = xr.open_zarr(store=store, consolidated=True)
elif ds_storage == 'local':
pooch.retrieve(
url="doi:10.5281/zenodo.7248056/hokkaido_japan.zip",
known_hash="md5:699b94e827c72bcd69bd786e56bfe5dc",
processor=pooch.Unzip(extract_dir='data'),
path=f".",
)
ds = xr.open_zarr('data/' + ds_name + '.zarr')
return ds
# function to generate the before and after dataset using a given aggregation method using the time step length
def before_after_ds(ds_path, ds_storage, ba_vars, aggregation, timestep_length, event_start_date, event_end_date):
ds = open_zarr_source(ds_path, ds_storage)
for var in ba_vars:
ds[var] = np.log(ds[var])
ds = ds.where(ds['sat:orbit_state'].compute() == 'd', drop=True)
before_ds = ds.drop_dims('timepair').sel(timestep=slice(None, event_start_date))
after_ds = ds.drop_dims('timepair').sel(timestep=slice(event_end_date, None))
if timestep_length < len(before_ds['timestep']):
before_ds = before_ds.isel(timestep=range(-1 - timestep_length, -1))
if timestep_length < len(after_ds['timestep']):
after_ds = after_ds.isel(timestep=range(timestep_length))
if aggregation == 'mean':
before_ds = before_ds.mean(dim=('timestep'))
after_ds = after_ds.mean(dim=('timestep'))
elif aggregation == 'median':
before_ds = before_ds.median(dim=('timestep'))
after_ds = after_ds.median(dim=('timestep'))
before_after_vars = []
for suffix in ['before', 'after']:
for var in ba_vars:
before_after_vars.append(f'{var}_{suffix}')
the_ds = before_ds.rename_vars({var: f'{var}_before' for var in ba_vars})
for var in ba_vars:
the_ds[f'{var}_after'] = after_ds[var]
for var in the_ds.data_vars:
the_ds[f'{var}_mean'] = the_ds[var].mean()
the_ds[f'{var}_std'] = the_ds[var].std()
return the_ds.compute().load()
# function to generate batches of data using xbatcher
def batching_dataset(ds, patch_size, input_vars, target, include_negatives):
mean_std_dict = {}
for var in input_vars:
if not mean_std_dict.get(var):
mean_std_dict[var] = {}
mean_std_dict[var]['mean'] = ds[f'{var}_mean'].values
mean_std_dict[var]['std'] = ds[f'{var}_std'].values
batches = []
bgen = xbatcher.BatchGenerator(ds, {'x': patch_size[0], 'y': patch_size[1]})
positives = 0
negatives = 0
for batch in bgen:
positives_tmp = batch[target].sum().item()
if not include_negatives and positives_tmp > 0:
positives = positives + positives_tmp
negatives += batch[target].size
batches.append(batch)
elif include_negatives and (batch['dem'] <= 0).sum() == 0:
positives = positives + positives_tmp
negatives += batch[target].size
batches.append(batch)
print(f"P/(P+N)", positives / negatives)
return batches, mean_std_dict
# class using the pytorch Dataset module to define the dataset
class BeforeAfterDatasetBatches(Dataset):
def __init__(self, batches, input_vars, target, mean_std_dict=None):
print("**************** INIT CALLED ******************")
self.batches = batches
self.target = target
self.input_vars = input_vars
self.mean = np.stack([mean_std_dict[var]['mean'] for var in input_vars]).reshape((-1, 1, 1))
self.std = np.stack([mean_std_dict[var]['std'] for var in input_vars]).reshape((-1, 1, 1))
def __len__(self):
return len(self.batches)
def __getitem__(self, idx):
batch = self.batches[idx]
inputs = np.stack([batch[var].values for var in self.input_vars])
inputs = (inputs - self.mean) / self.std
target = batch[self.target].values
inputs = np.nan_to_num(inputs, nan=0)
target = np.nan_to_num(target, nan=0)
target = (target > 0)
return inputs, target
# class using the lightning DataModule to define the data module implementing five key methods/steps
class BeforeAfterCubeDataModule(LightningDataModule):
"""LightningDataModule.
A DataModule implements 5 key methods:
- prepare_data (things to do on 1 GPU/TPU, not on every GPU/TPU in distributed mode)
- setup (things to do on every accelerator in distributed mode)
- train_dataloader (the training dataloader)
- val_dataloader (the validation dataloader(s))
- test_dataloader (the test dataloader(s))
This allows you to share a full dataset without explaining how to download,
split, transform and process the data.
Read the docs:
https://pytorch-lightning.readthedocs.io/en/latest/extensions/datamodules.html
"""
def __init__(
self,
ds_name: str,
ds_storage: str,
ba_vars,
aggregation,
timestep_length,
event_start_date,
event_end_date,
input_vars,
target,
include_negatives=False,
train_val_test_split: Tuple[float, float, float] = (0.7, 0.2, 0.1),
patch_size: Tuple[int, int] = (128, 128),
batch_size: int = 64,
num_workers: int = 0,
pin_memory: bool = False,
):
super().__init__()
# this line allows to access init params with 'self.hparams' attribute
self.save_hyperparameters(logger=False)
self.ds = None
self.batches = None
self.mean_std_dict = None
self.data_train: Optional[Dataset] = None
self.data_val: Optional[Dataset] = None
self.data_test: Optional[Dataset] = None
self.data_whole: Optional[Dataset] = None
def setup(self, stage: Optional[str] = None):
"""Load data. Set variables: `self.data_train`, `self.data_val`, `self.data_test`.
This method is called by lightning when doing `trainer.fit()` and `trainer.test()`,
so be careful not to execute the random split twice! The `stage` can be used to
differentiate whether it's called before trainer.fit()` or `trainer.test()`.
"""
# load datasets only if they're not loaded already
if not self.data_train and not self.data_val and not self.data_test:
self.ds = before_after_ds(self.hparams.ds_name, self.hparams.ds_storage, self.hparams.ba_vars, self.hparams.aggregation,
self.hparams.timestep_length, self.hparams.event_start_date,
self.hparams.event_end_date)
self.batches, self.mean_std_dict = batching_dataset(self.ds, self.hparams.patch_size, self.hparams.input_vars, self.hparams.target,
self.hparams.include_negatives)
dataset = BeforeAfterDatasetBatches(self.batches, self.hparams.input_vars, self.hparams.target,
mean_std_dict=self.mean_std_dict)
self.data_whole = dataset
if self.hparams.train_val_test_split:
train_val_test_split = [int(len(dataset) * x) for x in self.hparams.train_val_test_split]
train_val_test_split[2] = len(dataset) - train_val_test_split[1] - train_val_test_split[0]
train_val_test_split = tuple(train_val_test_split)
print("*" * 20)
print("Train - Val - Test SPLIT", train_val_test_split)
print("*" * 20)
self.data_train, self.data_val, self.data_test = random_split(
dataset=dataset,
lengths=train_val_test_split,
generator=torch.Generator().manual_seed(42),
)
print("*" * 20)
print("Train - Val - Test LENGTHS", len(self.data_train), len(self.data_val), len(self.data_test))
print("*" * 20)
def train_dataloader(self):
return MultiEpochsDataLoader(
dataset=self.data_train,
batch_size=self.hparams.batch_size,
num_workers=self.hparams.num_workers,
pin_memory=self.hparams.pin_memory,
shuffle=True,
persistent_workers=(self.hparams.num_workers > 0)
)
def val_dataloader(self):
return MultiEpochsDataLoader(
dataset=self.data_val,
batch_size=self.hparams.batch_size,
num_workers=self.hparams.num_workers,
pin_memory=self.hparams.pin_memory,
shuffle=False,
persistent_workers=(self.hparams.num_workers > 0)
)
def test_dataloader(self):
return MultiEpochsDataLoader(
dataset=self.data_test,
batch_size=self.hparams.batch_size,
num_workers=self.hparams.num_workers,
pin_memory=self.hparams.pin_memory,
shuffle=False,
persistent_workers=(self.hparams.num_workers > 0)
)
class MultiEpochsDataLoader(torch.utils.data.DataLoader):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self._DataLoader__initialized = False
self.batch_sampler = _RepeatSampler(self.batch_sampler)
self._DataLoader__initialized = True
self.iterator = super().__iter__()
def __len__(self):
return len(self.batch_sampler.sampler)
def __iter__(self):
for i in range(len(self)):
yield next(self.iterator)
class _RepeatSampler(object):
""" Sampler that repeats forever.
Args:
sampler (Sampler)
"""
def __init__(self, sampler):
self.sampler = sampler
def __iter__(self):
while True:
yield from iter(self.sampler)
U-Net Model#
The class below defines the U-Net architecture using pl.LightningModule.
Note
The U-Net model [RFB15]. is a convolutional neural network that was originally developed for biomedical image segmentation and extended to other applications. U-Net has an encoder-decoder architecture with skip connections that allow the model to capture both local and global features. The model is composed of a contracting path that captures context and a symmetric expanding path that enables precise localization.
For the landslide example, a RestNet50 (48,982,754 trainable parameters) is used as the encoder in the U-Net model. The model is trained using the Dice loss function and evaluated using the Area Under the Receiver Operating Characteristic (AUROC), Average Precision (AUPRC), and F1 score metrics.
class plUNET(pl.LightningModule):
def __init__(
self,
lr: float = 0.001,
weight_decay: float = 0.0005,
num_channels: int = 1,
loss='dice'
):
super().__init__()
self.save_hyperparameters(logger=False)
self.net = smp.UnetPlusPlus(encoder_name='resnet50', in_channels=num_channels, classes=2)
if loss == 'dice':
self.criterion = smp.losses.DiceLoss(mode='multiclass')
elif loss == 'ce':
self.criterion = torch.nn.CrossEntropyLoss()
self.train_auc = AUROC(task="binary")
self.train_f1 = F1Score(task="binary")
self.train_auprc = AveragePrecision(task="binary")
self.val_auc = AUROC(task="binary")
self.val_f1 = F1Score(task="binary")
self.val_auprc = AveragePrecision(task="binary")
self.test_auc = AUROC(task="binary")
self.test_auprc = AveragePrecision(task="binary")
self.test_f1 = F1Score(task="binary")
self.train_positive_count = 0
self.val_positive_count = 0
self.test_positive_count = 0
self.train_negative_count = 0
self.val_negative_count = 0
self.test_negative_count = 0
def forward(self, x: torch.Tensor):
return self.net(x)
def step(self, batch: Any):
x, y = batch
y = y.long()
logits = self.forward(x)
loss = self.criterion(logits, y)
preds = torch.nn.functional.softmax(logits, dim=1)[:, 1]
return loss, preds, y, x
def training_step(self, batch: Any, batch_idx: int):
loss, preds, targets, inputs = self.step(batch)
self.train_auc.update(preds, targets)
self.train_auprc.update(preds.flatten(), targets.flatten())
self.train_f1.update(preds.flatten(), targets.flatten())
self.log("train/loss", loss, on_step=False, on_epoch=True, prog_bar=True)
self.log("train/auc", self.train_auc, on_step=False, on_epoch=True, prog_bar=False)
self.log("train/auprc", self.train_auprc, on_step=False, on_epoch=True, prog_bar=False)
self.log("train/f1", self.train_f1, on_step=False, on_epoch=True, prog_bar=False)
return {"loss": loss, "preds": preds, "targets": targets, "inputs": inputs}
def on_train_epoch_end(self):
pass
def validation_step(self, batch: Any, batch_idx: int):
loss, preds, targets, _ = self.step(batch)
# log val metrics
self.val_auc.update(preds, targets)
self.val_auprc.update(preds.flatten(), targets.flatten())
self.val_f1.update(preds.flatten(), targets.flatten())
self.log("val/loss", loss, on_step=False, on_epoch=True, prog_bar=True)
self.log("val/auc", self.val_auc, on_step=False, on_epoch=True, prog_bar=True)
self.log("val/auprc", self.val_auprc, on_step=False, on_epoch=True, prog_bar=True)
self.log("val/f1", self.val_f1, on_step=False, on_epoch=True, prog_bar=True)
return {"loss": loss, "preds": preds, "targets": targets}
def test_step(self, batch: Any, batch_idx: int):
loss, preds, targets, _ = self.step(batch)
self.test_auc.update(preds, targets)
self.test_auprc.update(preds.flatten(), targets.flatten())
self.test_f1.update(preds.flatten(), targets.flatten())
self.log("test/loss", loss, on_step=False, on_epoch=True)
self.log("test/auc", self.test_auc, on_step=False, on_epoch=True, prog_bar=False)
self.log("test/auprc", self.test_auprc, on_step=False, on_epoch=True, prog_bar=False)
self.log("test/f1", self.test_f1, on_step=False, on_epoch=True, prog_bar=False)
return {"loss": loss, "preds": preds, "targets": targets}
def on_test_epoch_end(self):
pass
def configure_optimizers(self):
optimizer = torch.optim.Adam(
params=self.parameters(), lr=self.hparams.lr, weight_decay=self.hparams.weight_decay
)
lr_scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer)
return {'optimizer': optimizer, 'lr_scheduler': lr_scheduler, "monitor": "train/loss"}
def save(self, filename):
torch.save(self.state_dict(), filename)
def load(self, filename):
self.load_state_dict(torch.load(filename))
Pipeline#
The pipeline consists in defining and initiating the datacube, defining and initiating the U-Net model, training the model, saving the trained model. Finally, we evaluate the model using the test dataset and visualize the results over one of the test batches.
Set a seed (reproducibility)#
seed_everything(42, workers=True)
Seed set to 42
42
Define the datacube#
hparams["input_vars"] = hparams["input_vars"].split(',')
dm = BeforeAfterCubeDataModule(
ds_name=hparams["ds_name"],
ds_storage=hparams["ds_storage"],
ba_vars=['vv', 'vh'],
aggregation='mean',
timestep_length=hparams["timestep_length"],
event_start_date='20180905',
event_end_date='20180907',
input_vars=hparams["input_vars"],
target='landslides',
train_val_test_split=(0.7, 0.2, 0.1),
patch_size=hparams["patch_size"],
batch_size=hparams["batch_size"],
include_negatives=False
)
Initiate the datacube#
dm.setup()
P/(P+N) 0.09151421704338593
**************** INIT CALLED ******************
********************
Train - Val - Test SPLIT (216, 61, 32)
********************
********************
Train - Val - Test LENGTHS 216 61 32
********************
U-Net Model#
Model training#
We start defining the model using the plUNET class and the hyperparameters defined in the hparams dictionary.
model = plUNET(
lr=hparams["lr"],
weight_decay=hparams["weight_decay"],
num_channels=len(hparams["input_vars"]),
loss=hparams["loss"]
)
Then we initiate the trainer with the maximum number of epochs defined in the hparams dictionary.
trainer = pl.Trainer(max_epochs=hparams["max_epochs"], accelerator="auto")
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
/srv/conda/envs/notebook/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/logger_connector/logger_connector.py:67: Starting from v1.9.0, `tensorboardX` has been removed as a dependency of the `pytorch_lightning` package, due to potential conflicts with other packages in the ML ecosystem. For this reason, `logger=True` will use `CSVLogger` as the default logger, unless the `tensorboard` or `tensorboardX` packages are found. Please `pip install lightning[extra]` or one of them to enable TensorBoard support by default
Finally, we fit the trainer with the model and the datacube.
trainer.fit(model, dm)
You are using a CUDA device ('NVIDIA A100-SXM4-40GB') that has Tensor Cores. To properly utilize them, you should set `torch.set_float32_matmul_precision('medium' | 'high')` which will trade-off precision for performance. For more details, read https://pytorch.org/docs/stable/generated/torch.set_float32_matmul_precision.html#torch.set_float32_matmul_precision
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
| Name | Type | Params
--------------------------------------------------------
0 | net | UnetPlusPlus | 49.0 M
1 | criterion | CrossEntropyLoss | 0
2 | train_auc | BinaryAUROC | 0
3 | train_f1 | BinaryF1Score | 0
4 | train_auprc | BinaryAveragePrecision | 0
5 | val_auc | BinaryAUROC | 0
6 | val_f1 | BinaryF1Score | 0
7 | val_auprc | BinaryAveragePrecision | 0
8 | test_auc | BinaryAUROC | 0
9 | test_auprc | BinaryAveragePrecision | 0
10 | test_f1 | BinaryF1Score | 0
--------------------------------------------------------
49.0 M Trainable params
0 Non-trainable params
49.0 M Total params
196.006 Total estimated model params size (MB)
/srv/conda/envs/notebook/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/data_connector.py:441: The 'val_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=254` in the `DataLoader` to improve performance.
/srv/conda/envs/notebook/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/data_connector.py:441: The 'train_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=254` in the `DataLoader` to improve performance.
/srv/conda/envs/notebook/lib/python3.11/site-packages/pytorch_lightning/loops/fit_loop.py:293: The number of training batches (4) is smaller than the logging interval Trainer(log_every_n_steps=50). Set a lower value for log_every_n_steps if you want to see logs for the training epoch.
`Trainer.fit` stopped: `max_epochs=30` reached.
Save the trained model#
model.save("landslide.pt")
Model Evaluation#
We assess the model performance using the test dataset and the best model checkpoint.
result = trainer.test(ckpt_path="best", datamodule=dm)
Restoring states from the checkpoint path at /home/jovyan/igarss/repos/pangeo-igarss2024/docs/lightning_logs/version_25/checkpoints/epoch=29-step=120.ckpt
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Loaded model weights from the checkpoint at /home/jovyan/igarss/repos/pangeo-igarss2024/docs/lightning_logs/version_25/checkpoints/epoch=29-step=120.ckpt
/srv/conda/envs/notebook/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/data_connector.py:441: The 'test_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=254` in the `DataLoader` to improve performance.
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Test metric ┃ DataLoader 0 ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ test/auc │ 0.8914892673492432 │ │ test/auprc │ 0.5419434309005737 │ │ test/f1 │ 0.04432787001132965 │ │ test/loss │ 0.24052730202674866 │ └───────────────────────────┴───────────────────────────┘
def visualise(inputs, prediction, target):
npinputs = inputs.detach().cpu().numpy()
npprediction = prediction.detach().cpu().numpy()
nptarget = target.detach().cpu().numpy()
f, axarr = plt.subplots(1,10, figsize=(25,8))
axarr[0].imshow(npprediction)
axarr[0].set_title('Prediction', size='x-large')
axarr[1].imshow(nptarget)
axarr[1].set_title('Truth', size='x-large')
for i in range(len(hparams["input_vars"])):
axarr[i+2].imshow(npinputs[i])
axarr[i+2].set_title(hparams["input_vars"][i], size='x-large')
# get some random training images
dataiter = iter(dm.test_dataloader())
images, target = next(dataiter)
prediction = model(images).argmax(dim=1)
batchid = 0
visualise(images[batchid],target[batchid],prediction[batchid])
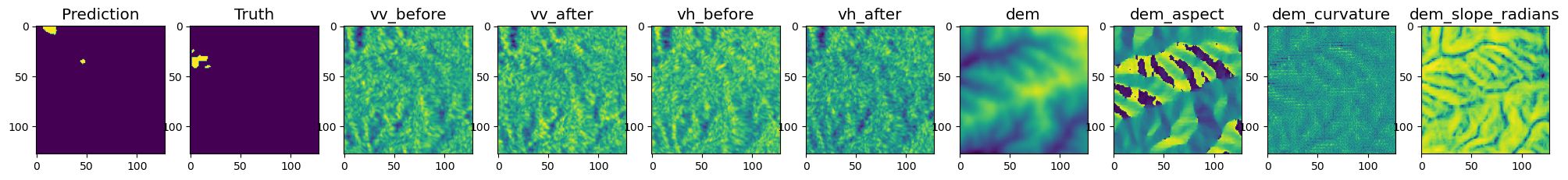
Prediction over larger batches#
Fully convolutional networks like U-Net do not have fixed input shape, so we could try a different size input for the trained model. We demonstrate how to load the trained model and evaluate it over a larger batch of 1024 pixels x 1024 pixels of data.
Load the trained model#
The function below calls the saved file with the trained model from a remote location or local file.
def get_state_dict(weight_fpath: str, device: torch.device):
if "s3://" in weight_fpath:
s3 = s3fs.S3FileSystem(anon=True,
client_kwargs={
"endpoint_url": "https://pangeo-eosc-minioapi.vm.fedcloud.eu/"
})
with s3.open(weight_fpath, "rb") as f:
buffer = io.BytesIO(f.read())
state_dict = torch.load(buffer, map_location=device)
else:
state_dict = torch.load(f=weight_fpath, map_location=device)
return state_dict
We define the model and load the trained weights from a remote location.
model_path = "s3://acocacastro2-igarss24/landslide.pt" #change to landslide.pt to load the local file
model = plUNET(
num_channels=len(hparams["input_vars"]))
state_dict = get_state_dict(model_path,'cuda')
model.load_state_dict(state_dict)
model.eval()
Show code cell output
plUNET(
(net): UnetPlusPlus(
(encoder): ResNetEncoder(
(conv1): Conv2d(8, 64, kernel_size=(7, 7), stride=(2, 2), padding=(3, 3), bias=False)
(bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(maxpool): MaxPool2d(kernel_size=3, stride=2, padding=1, dilation=1, ceil_mode=False)
(layer1): Sequential(
(0): Bottleneck(
(conv1): Conv2d(64, 64, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(64, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(downsample): Sequential(
(0): Conv2d(64, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): Bottleneck(
(conv1): Conv2d(256, 64, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(64, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
(2): Bottleneck(
(conv1): Conv2d(256, 64, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(64, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
)
(layer2): Sequential(
(0): Bottleneck(
(conv1): Conv2d(256, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(128, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(downsample): Sequential(
(0): Conv2d(256, 512, kernel_size=(1, 1), stride=(2, 2), bias=False)
(1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): Bottleneck(
(conv1): Conv2d(512, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(128, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
(2): Bottleneck(
(conv1): Conv2d(512, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(128, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
(3): Bottleneck(
(conv1): Conv2d(512, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(128, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
)
(layer3): Sequential(
(0): Bottleneck(
(conv1): Conv2d(512, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(256, 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(downsample): Sequential(
(0): Conv2d(512, 1024, kernel_size=(1, 1), stride=(2, 2), bias=False)
(1): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): Bottleneck(
(conv1): Conv2d(1024, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(256, 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
(2): Bottleneck(
(conv1): Conv2d(1024, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(256, 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
(3): Bottleneck(
(conv1): Conv2d(1024, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(256, 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
(4): Bottleneck(
(conv1): Conv2d(1024, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(256, 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
(5): Bottleneck(
(conv1): Conv2d(1024, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(256, 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
)
(layer4): Sequential(
(0): Bottleneck(
(conv1): Conv2d(1024, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(512, 512, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(512, 2048, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(2048, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(downsample): Sequential(
(0): Conv2d(1024, 2048, kernel_size=(1, 1), stride=(2, 2), bias=False)
(1): BatchNorm2d(2048, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): Bottleneck(
(conv1): Conv2d(2048, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(512, 2048, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(2048, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
(2): Bottleneck(
(conv1): Conv2d(2048, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv3): Conv2d(512, 2048, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): BatchNorm2d(2048, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
)
)
)
(decoder): UnetPlusPlusDecoder(
(center): Identity()
(blocks): ModuleDict(
(x_0_0): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(3072, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_0_1): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(1280, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_1_1): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(1536, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_0_2): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(896, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_1_2): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(1024, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_2_2): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(768, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_0_3): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(320, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(32, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(32, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(32, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_1_3): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(448, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_2_3): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(384, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_3_3): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(320, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
(x_0_4): DecoderBlock(
(conv1): Conv2dReLU(
(0): Conv2d(32, 16, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention1): Attention(
(attention): Identity()
)
(conv2): Conv2dReLU(
(0): Conv2d(16, 16, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU(inplace=True)
)
(attention2): Attention(
(attention): Identity()
)
)
)
)
(segmentation_head): SegmentationHead(
(0): Conv2d(16, 2, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(1): Identity()
(2): Activation(
(activation): Identity()
)
)
)
(criterion): DiceLoss()
(train_auc): BinaryAUROC()
(train_f1): BinaryF1Score()
(train_auprc): BinaryAveragePrecision()
(val_auc): BinaryAUROC()
(val_f1): BinaryF1Score()
(val_auprc): BinaryAveragePrecision()
(test_auc): BinaryAUROC()
(test_auprc): BinaryAveragePrecision()
(test_f1): BinaryF1Score()
)
Set the experiment#
We copy the original hyperparameters and change the patch size to 1024x1024 pixels and the batch size to 1.
hparams_all = copy.deepcopy(hparams)
hparams_all['patch_size']=[1024,1024]
hparams_all['batch_size']=1
The original data module is initiated data that discarded batches with negative values (no landslides). We set include_negatives=True to include all batches and predict over the whole surface.
dm_all= BeforeAfterCubeDataModule(
ds_name=hparams["ds_name"],
ds_storage=hparams["ds_storage"],
ba_vars=['vv', 'vh'],
aggregation='mean',
timestep_length=hparams["timestep_length"],
event_start_date='20180905',
event_end_date='20180907',
input_vars=hparams["input_vars"],
target='landslides',
train_val_test_split=None,
patch_size=hparams_all["patch_size"],
batch_size=hparams_all["batch_size"],
include_negatives=True
)
dm_all.setup()
P/(P+N) 0.022092103958129883
**************** INIT CALLED ******************
We set a dataloader to load the whole dataset.
ds = dm_all.data_whole
dataloader = torch.utils.data.DataLoader(ds, pin_memory=False, shuffle=False)
Model inference#
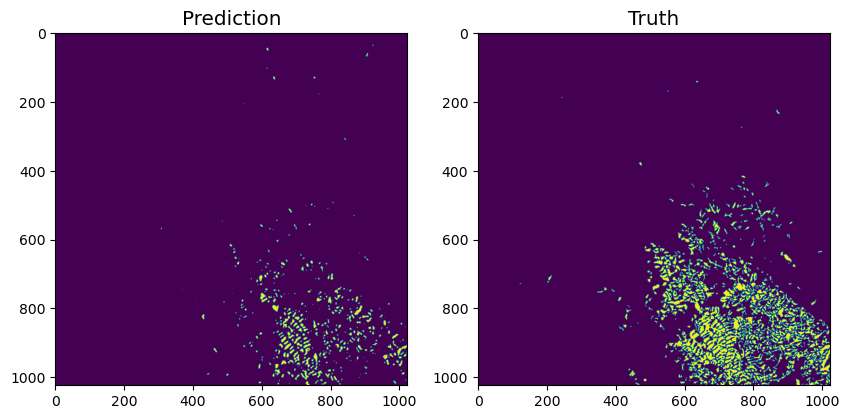
We run the model inference over the whole dataset and visualise over a batch of 1024 pixels x 1024 pixels.
targets = []
outputs = []
for batch_idx, (image, target) in tqdm(enumerate(dataloader), total=len(dataloader)):
output = model(image)
outputs.append(output[:, 0].cpu().detach().numpy())
targets.append(target.cpu().detach().numpy())
targets = np.concatenate(targets, axis=0)
outputs = np.concatenate(outputs, axis=0)
batch_id = 6
f, axarr = plt.subplots(1,2, figsize=(10,10))
axarr[0].imshow(outputs[batch_id]<0.5)
axarr[0].set_title('Prediction', size='x-large')
axarr[1].imshow(targets[batch_id])
axarr[1].set_title('Truth', size='x-large')
plt.show()
Visualise results using the HoloViz Ecosystem#
The cells below demonstrate how to use the HoloViz ecosystem that it is compatible with xarray.
import holoviews as hv
import panel as pn
import hvplot.xarray
import random
random.seed(42)
hv.extension('bokeh')
pn.extension()
We will create a dashboard to interactively display predictions and truth numpy arrays (transformed to xarray). The dashboard includes a widget to explore the available batches (Total=20). For the purpose of this notebook will only show 5 randomly sampled batches to export the dashboard using an embedding state.
general_settings = {'x':'x', 'y':'y', 'data_aspect':1, 'flip_yaxis':True,
'xaxis':False, 'yaxis':None, 'tools':['tap', 'box_select'], 'cmap':'viridis', 'colorbar':False}
# set widgets
batch_list = list(random.sample(range(0, len(dataloader)), 5)) #select 5 randomly batches
#batch_list = list(range(0, len(dataloader)) #uncomment to pass all available batches
target_batch = pn.widgets.Select(name = 'Batch ID', options = batch_list)
@pn.depends(target_batch.param.value)
def plot_visualise(batch_id):
prediction_xr = xr.DataArray(outputs[batch_id]<0.5, dims=['y', 'x'],
coords={'y': np.arange(outputs[batch_id].shape[0]),
'x': np.arange(outputs[batch_id].shape[1])})
prediction = prediction_xr.hvplot(**general_settings, title='Prediction')
target_xr = xr.DataArray(targets[batch_id], dims=['y', 'x'],
coords={'y': np.arange(outputs[batch_id].shape[0]),
'x': np.arange(outputs[batch_id].shape[1])})
target = target_xr.hvplot(**general_settings, title='Truth')
p = prediction + target
return p
plot_batches = pn.Row(
plot_visualise,
pn.Column(pn.Spacer(height=5), target_batch, styles={'background':'#f0f0f0'}, sizing_mode="fixed"),
width_policy='max', height_policy='max',
)
plot_batches.embed()


