Data access and discovery
Contents
Data access and discovery#
Context#
We will be using Long Term Statistics (1999-2019) product provided by the Copernicus Global Land Service over Lombardia and access them through S3-compatible storage. We will also explore Sentinel-2 Cloud-Optimised Dataset online through SpatioTemporal Asset Catalogs (STAC).
Setup#
This episode uses the following main Python packages:
s3fs [S3FsDTeam16]
Please install these packages if not already available in your Python environment.
Packages#
In this episode, Python packages are imported when we start to use them. However, for best software practices, we recommend you to install and import all the necessary libraries at the top of your Jupyter notebook.
Introduction to the Long Term statistics#
CGLS LTS are computed over a time span of 20 years aggregated over separate 10-day periods (month/01,month/11, month/21). For each date the long term minimum, maximum, mean, median and standard deviation are computed.
S3-compatible Object Storage to access online data#
Up to now we have downloaded data locally and then opened with Xarray open_dataset. When willing to manipulate large amount of data, this approach is not optimal (since it requires a lot of unnecessary local downloads). Sharing data online as Object Storage allows for data sharing and access to much larger amounts of data.
One of the most popular methods to access online remote data is through Amazon Simple Storage Service (S3) and you don’t necessarily need to use Amazon services to benefit from S3 object storage. Many other providers offer S3-compatible object storage that can be accessed in a very similar way.
Below we will be accessing online the NDVI Long Term Statistics from Copernicus Land Service that we have publicly stored in OpenStack Object storage (Swift).
import s3fs
import xarray as xr
fs = s3fs.S3FileSystem(anon=True,
client_kwargs={
'endpoint_url': 'https://object-store.cloud.muni.cz'
})
Tip
The parameter anon is for anonymous and is set to True because the data we have stored at https://object-store.cloud.muni.cz is public
List files and folders in existing buckets#
Instead of organizing files in various folders, object storage systems store files in a flat organization of containers (called “buckets”).
fs.ls('foss4g-data')
['foss4g-data/CGLS_LTS_1999_2019',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia',
'foss4g-data/test']
fs.ls('foss4g-data/CGLS_LTS_1999_2019_Lombardia')
['foss4g-data/CGLS_LTS_1999_2019_Lombardia/',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0101_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0111_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0121_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0201_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0211_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0221_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0301_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0311_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0321_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0401_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0411_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0421_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0501_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0511_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0521_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0601_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0611_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0621_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0701_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0711_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0721_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0801_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0811_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0821_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0901_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0911_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0921_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1001_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1011_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1021_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1101_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1111_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1121_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1201_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1211_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1221_GLOBE_VGT-PROBAV_V3.0.1.nc']
Access remote files from S3-compatible Object Storage#
s3path = 's3://foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0101_GLOBE_VGT-PROBAV_V3.0.1.nc'
LTS = xr.open_dataset(fs.open(s3path))
LTS
<xarray.Dataset>
Dimensions: (lon: 331, lat: 235)
Coordinates:
* lon (lon) float64 8.5 8.509 8.518 8.527 ... 11.42 11.43 11.44 11.45
* lat (lat) float64 46.7 46.69 46.68 46.67 ... 44.63 44.63 44.62 44.61
Data variables:
crs |S1 b''
min (lat, lon) float32 nan nan -0.076 -0.048 ... 0.216 0.256 0.236
median (lat, lon) float32 nan nan -0.072 -0.048 ... 0.404 0.404 0.4 0.416
max (lat, lon) float32 nan nan -0.028 -0.048 ... 0.56 0.54 0.568 0.588
mean (lat, lon) float32 nan nan -0.064 -0.048 ... 0.388 0.392 0.404 0.42
stdev (lat, lon) float32 nan nan 0.024 nan ... 0.088 0.088 0.088 0.104
nobs (lat, lon) float32 nan nan 4.0 1.0 nan ... 21.0 21.0 21.0 21.0 21.0
Attributes: (12/19)
Conventions: CF-1.6
parent_identifier: urn:cgls:global:ndvi_stats_all
identifier: urn:cgls:global:ndvi_stats_all:NDVI-LTS_1999-2019-0...
long_name: Normalized Difference Vegetation Index
title: Normalized Difference Vegetation Index: Long Term S...
product_version: V3.0.1
... ...
source: Derived from EO satellite imagery
processing_mode: Offline
references: https://land.copernicus.eu/global/products/ndvi
copyright: Copernicus Service information 2021
archive_facility: VITO
history: 2021-03-01 - Processing line NDVI LTSLTS.sel(lat=45.88, lon=8.63, method='nearest')['min'].values
array(0.264, dtype=float32)
Warning
The same dataset can be available from different locations e.g. CGLS distributor VITO, Zenodo, S3-compatible OpenStack Object storage (Swift), etc. How do you know if it corresponds to the very same dataset? You cannot know except if the datasets have a persistent identifier such as a Digital Object Identifier. It is therefore recommended 1) to be extra careful about where you get your datasets, and 2) to double check that the content is exactly what you expect (for instance, you can perform basic quality checks).
Access multiple remote files#
s3path = 's3://foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_*.nc'
remote_files = fs.glob(s3path)
remote_files
['foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0101_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0111_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0121_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0201_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0211_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0221_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0301_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0311_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0321_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0401_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0411_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0421_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0501_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0511_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0521_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0601_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0611_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0621_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0701_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0711_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0721_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0801_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0811_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0821_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0901_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0911_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-0921_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1001_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1011_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1021_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1101_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1111_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1121_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1201_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1211_GLOBE_VGT-PROBAV_V3.0.1.nc',
'foss4g-data/CGLS_LTS_1999_2019_Lombardia/AOI_c_gls_NDVI-LTS_1999-2019-1221_GLOBE_VGT-PROBAV_V3.0.1.nc']
We need to add a time dimension to concatenate data. For this, we define a function that will be called for each remote file (via the preprocess parameter of Xarray open_mfdataset.)
from datetime import datetime
def preprocess(ds):
t = datetime.strptime(ds.attrs['identifier'].split(':')[-1].split('_')[1].replace('1999-', ''), "%Y-%m%d")
return(ds.assign_coords(time=t).expand_dims('time'))
Xarray open_mfdataset allows opening multiple files at the same time.
# Iterate through remote_files to create a fileset
fileset = [fs.open(file) for file in remote_files]
When opening remote files, you can also select the variables you wish to analyze.
LTS = xr.open_mfdataset(fileset, combine='nested', concat_dim=['time'], preprocess=preprocess,
decode_coords="all")
LTS
<xarray.Dataset>
Dimensions: (lon: 331, lat: 235, time: 36)
Coordinates:
* lon (lon) float64 8.5 8.509 8.518 8.527 ... 11.42 11.43 11.44 11.45
* lat (lat) float64 46.7 46.69 46.68 46.67 ... 44.63 44.63 44.62 44.61
crs |S1 b''
* time (time) datetime64[ns] 2019-01-01 2019-01-11 ... 2019-12-21
Data variables:
min (time, lat, lon) float32 dask.array<chunksize=(1, 235, 331), meta=np.ndarray>
median (time, lat, lon) float32 dask.array<chunksize=(1, 235, 331), meta=np.ndarray>
max (time, lat, lon) float32 dask.array<chunksize=(1, 235, 331), meta=np.ndarray>
mean (time, lat, lon) float32 dask.array<chunksize=(1, 235, 331), meta=np.ndarray>
stdev (time, lat, lon) float32 dask.array<chunksize=(1, 235, 331), meta=np.ndarray>
nobs (time, lat, lon) float32 dask.array<chunksize=(1, 235, 331), meta=np.ndarray>
Attributes: (12/19)
Conventions: CF-1.6
parent_identifier: urn:cgls:global:ndvi_stats_all
identifier: urn:cgls:global:ndvi_stats_all:NDVI-LTS_1999-2019-0...
long_name: Normalized Difference Vegetation Index
title: Normalized Difference Vegetation Index: Long Term S...
product_version: V3.0.1
... ...
source: Derived from EO satellite imagery
processing_mode: Offline
references: https://land.copernicus.eu/global/products/ndvi
copyright: Copernicus Service information 2021
archive_facility: VITO
history: 2021-03-01 - Processing line NDVI LTSTip
If you use one of xarray’s open methods such as xarray.open_dataset to load netCDF files with the default engine, it is recommended to use decode_coords=”all”. This will load the grid mapping variable into coordinates for compatibility with rioxarray. See rioxarray documentation.
Preparing and discover online datasets#
With the plethora of cloud storage, there are many available online datasets. To ease the preparation and discovery of such datasets, we describe emerging community-driven initiatives promoting standards suited to both geospatial and geoscience communities. Most of the material below is adapted from a previous Pangeo 101 training [GAF2)].
Tip
While we provide a general intro to some initiatives, we suggest below a list of FOSS4G 2022 talks with very interesting developments to prepare and discover spatio-temporal datasets in the cloud. Enjoy!
STAC Best Practices and Tools, 2022-08-24, 11:00–11:30
Early use of FOSS4G in a space start up, 2022-08-24, 11:30–12:00
Exploring Data Interoperability with STAC and the Microsoft Planetary Computer, 2022-08-24, 12:10–12:15
Serving oblique aerial imagery using STAC and Cloud Optimized Geotiffs, 2022-08-24, 14:45–15:15
Pangeo Forge: Crowdsourcing Open Data in the Cloud. 2022-08-26, 10:00-10:30.
Analysis Ready, cloud optimized data (ARCO)#
When analyzing data at scale, the data format used is key. For years, the main data format was netCDF e.g. Network Common Data Form but with the use of cloud computing and interest in Open Science, different formats are often more suitable.
Formats for analyzing data from the cloud are refered to as “Analysis Ready, Cloud Optimized” data formats or in short ARCO. Find further info about ARCO datasets in [SAH+22].
What is “Analysis Ready”?
Think in terms of “Datasets” not “data files”
No need for tedious homogenizing / cleaning setup guides
Curated and cataloged
What is “Cloud Optimized”?
Compatible with object storage e.g. access via HTTP
Supports lazy access and intelligent subsetting
Integrates with high-level analysis libraries and distributed frameworks
Instead of having a big dataset, ARCO datasets are chunked appropriately for analysis and have rich metadata (See Figure 1).

Fig 1. Example of an ARCO dataset. Source: [GAF2)].
The Pangeo forge initiative#
Pangeo Forge is an open source platform for data Extraction, Transformation, and Loading (ETL). The goal of Pangeo Forge is to make it easy to extract data from traditional repositories and deposit this data in cloud object storage in an analysis-ready, cloud optimized (ARCO) format [GAF2)].
Pangeo Forge is inspired directly by Conda Forge, a community-led collection of recipes for building conda packages.
It is under active development and the Pangeo community hopes it will play a role in democratizing the publication of datasets in ARCO format.
How does Pangeo Forge work?#
Pangeo Forge defines the concept of a recipe, which specifies the logic for transforming a specific data archive into an ARCO data store. All contributions to Pangeo Forge must include an executable Python module, named recipe.py or similar, in which the data transformation logic is embedded (Figure 2). The recipe contributor is expected to use one of a predefined set of template algorithms defined by Pangeo Forge. Each of these templated algorithms is designed to transform data of a particular source type into a corresponding ARCO format, and requires only that the contributor populate the template with information unique to their specific data transformation, including the location of the source files and the way in which they should be aligned in the resulting ARCO data store [SAH+22].
The diagram below looks complicated but like for conda forge most of the process is automated.
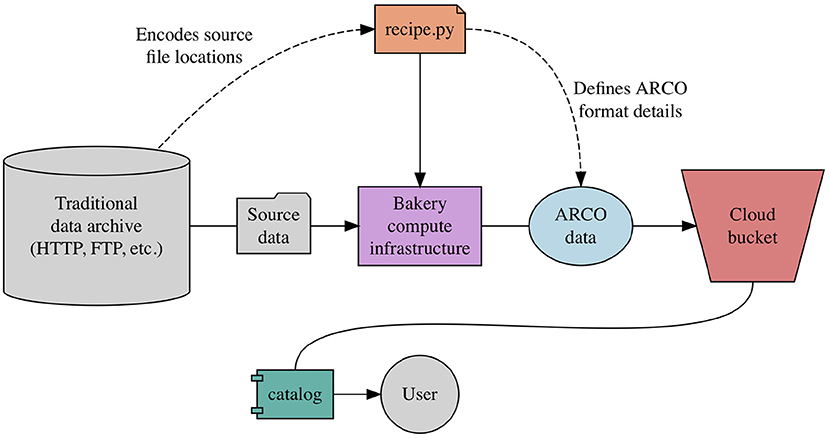
Fig 2. A recipe in relation to Pangeo Forge architecture. Source: [SAH+22].
The next step after preparing the dataset is then to tell the community where and how to access to your transformed dataset.
This is done by creating a catalog.
Spatio Temporal Asset Catalogs (STAC)#
The STAC specification is a common language to describe geospatial information, so it can more easily be worked with, indexed, and discovered.
Why STAC?#
Each provider has its own catalog and interface (APIs).
Every time you want to access a new catalog, you need to change your program.
We have lots of data providers and each with a bespoke interface.
It is becoming quickly difficult for programmers who need to design a new data connector each time.
Features#
STAC catalogs are extremely simple.
They are composed of three layers:
Catalogs
Collections
Items
STAC is very popular for Earth Observation satellite imagery.
For instance it can be used to access Sentinel-2 in AWS (see Figure 3).

Fig 3. Example of STAC collection of Sentinel-2 images hosted in AWS. Source: [GAF2)].
STAC and Pangeo Forge#
Pangeo-forge supports the creation of analysis-ready cloud optimized (ARCO) data in cloud object storage from “classical” data repositories.
STAC is used to create catalogs and goes beyond the Pangeo ecosystem.
Work is ongoing to figure out the best way to expose Pangeo-Forge-generated data assets via STAC catalogs.
Tip
Pangeo members, Scott Henderson (University of Washington) and Tom Augspurger (Microsoft), provided a great workshop in FOSS4G 2021 covering STAC.
Feel free to explore the GitHub repository of the here.
- Access to remote dataset
- ARCO datasets
- Pangeo Forge
- STAC
References#
- GAF2)(1,2,3,4)
Basile Goussard, Ryan Abernathey, and Anne Fouilloux. The pangeo ecosystem. https://training.galaxyproject.org/training-material/topics/climate/tutorials/pangeo-notebook/slides.html#1, 2022 (accessed August 7, 2022).
- SAH+22(1,2,3)
Charles Stern, Ryan Abernathey, Joseph Hamman, Rachel Wegener, Chiara Lepore, Sean Harkins, and Alexander Merose. Pangeo forge: crowdsourcing analysis-ready, cloud optimized data production. Frontiers in Climate, 2022. URL: https://www.frontiersin.org/articles/10.3389/fclim.2021.782909, doi:10.3389/fclim.2021.782909.
Packages citation#
- HH17
S. Hoyer and J. Hamman. Xarray: N-D labeled arrays and datasets in Python. Journal of Open Research Software, 2017. URL: https://doi.org/10.5334/jors.148, doi:10.5334/jors.148.
- S3FsDTeam16
S3Fs Development Team. S3Fs. 2016. URL: https://github.com/fsspec/s3fs/.


