Python Ecosystem for EO#
Core Python Libraries Overview#
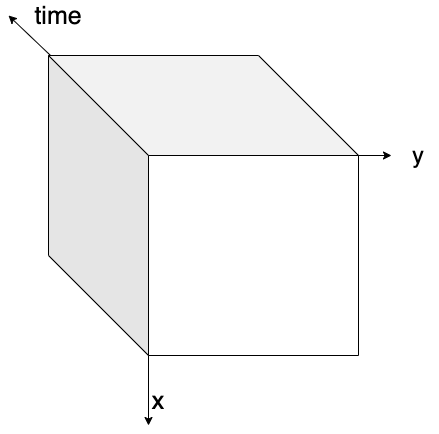
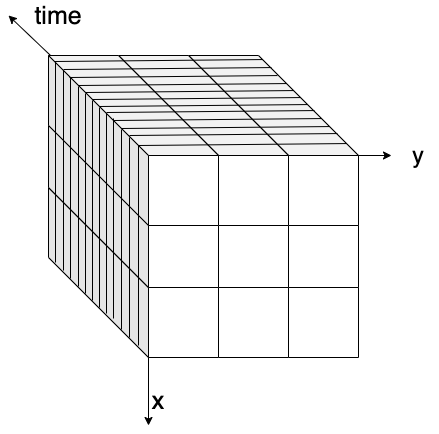
Python has become the lingua franca of EO processing through powerful libraries. Before diving into the details, let’s revisit data cubes and how we can represent them in Python. EO data cubes in Python are typically represented as labeled multidimensional arrays that integrate data values, coordinates, and metadata. Consider, for example, a 6x7 raster with 4 spectral bands collected over 3 time points. This structured representation allows efficient and intuitive access to complex EO datasets.
Fig. 1 An examplary raster datacube with 4 dimensions: x, y, bands and time.#
The demo below uses the Pangeo ecosystem to access and process data.
Note
The Pangeo ecosystem Pangeo is a Community-Driven Approach to Advancing Open Source Earth Observation Tools Across Disciplines.
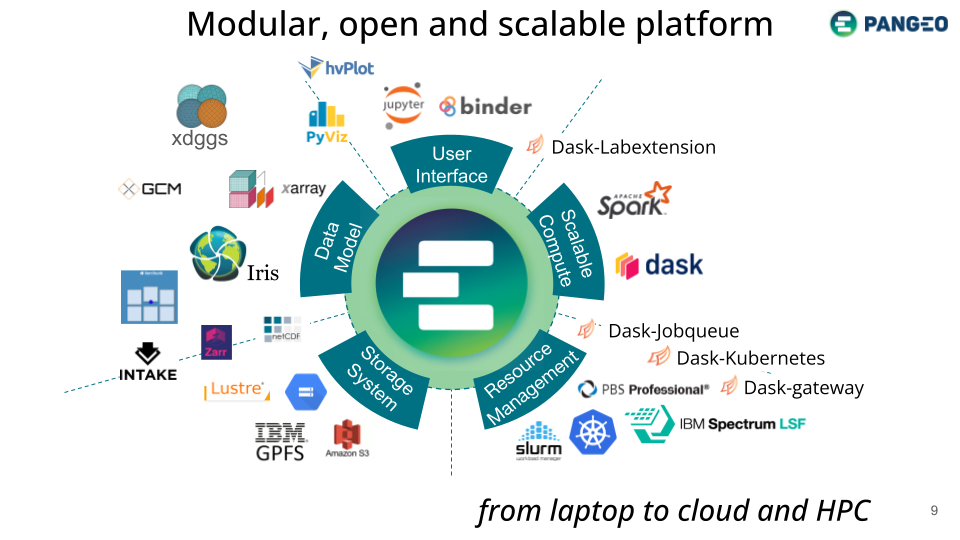
The list of Python packages below is not exhaustive. Its purpose is mainly to draw a visual picture of the different components typically needed for Earth Observation workflows.
Data Models#
xarray → N-dimensional arrays (gridded data).
geopandas → Tabular vector data (points, lines, polygons).
shapely → Geometries & spatial operations.
pyproj → Coordinate systems & reprojection.
eopf-xarray → Extends xarray with EO Processing Framework methods.
Storage Solutions (I/O & Formats)#
rioxarray → GeoTIFFs + CRS handling.
rasterio → Raster I/O.
fiona → Vector I/O (GeoJSON, Shapefile).
Zarr / NetCDF (via xarray) → Cloud-native, chunked array storage.
Catalogs & Metadata (optional)#
Catalogs are not strictly required — you can load files directly — but they become invaluable when dealing with large, multi-sensor, or cloud-hosted EO datasets.
pystac → STAC objects (Items, Collections, Catalogs).
sat-search → Search STAC APIs.
stackstac → Turn STAC items into xarray stacks.
odc-stac → Load STAC into xarray (optimized for EO).
Scalable Compute#
dask → Parallel/distributed computing.
pangeo stack → Dask + xarray ecosystem for scalable EO.
User Interfaces & Visualization#
hvplot → High-level, interactive plotting (works with xarray, geopandas, dask).
holoviews / panel → Dashboards and advanced UIs.
folium / ipyleaflet → Interactive maps in notebooks.
cartopy → Map projections & static plots.
Resource Management & Deployment (optional)#
Kubernetes / Dask Gateway → Manage compute clusters.
Pangeo Hub / JupyterHub → Multi-user access to scalable EO environments.
Data cubes and Lazy data loading with Xarray#
When accessing data through an API or cloud service, data is often lazily loaded. This means that initially only the metadata is retrieved, allowing us to understand the data’s dimensions, spatial and temporal extents, available spectral bands, and additional context without downloading the entire dataset.
Xarray supports this approach effectively, providing a powerful interface to open, explore, and manipulate large EO data cubes efficiently.
Let’s open an example dataset to explore these capabilities.
Note
xarray-eopf xarray-eopf is a Python package that extends xarray with a custom backend called “eopf-zarr”. This backend enables seamless access to ESA EOPF data products stored in the Zarr format, presenting them as analysis-ready data structures.
This notebook demonstrates how to use the xarray-eopf backend to explore and analyze EOPF Zarr datasets. It highlights the key features currently supported by the backend.
🐙 GitHub: EOPF Sample Service – xarray-eopf
📘 Documentation: xarray-eopf Docs
import xarray as xr
xr.set_options(display_expand_attrs=False)
<xarray.core.options.set_options at 0x1083ddb90>
path = (
"https://objects.eodc.eu/e05ab01a9d56408d82ac32d69a5aae2a:202505-s02msil2a/18/products"
"/cpm_v256/S2B_MSIL2A_20250518T112119_N0511_R037_T29RLL_20250518T140519.zarr"
)
ds = xr.open_datatree(path, engine="eopf-zarr", op_mode="native", chunks={})
ds
<xarray.DatasetView> Size: 0B
Dimensions: ()
Data variables:
*empty*
Attributes: (2)<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*conditions<xarray.DatasetView> Size: 559kB Dimensions: (angle: 2, band: 13, y: 23, x: 23, detector: 5) Coordinates: * angle (angle) <U7 56B 'zenith' 'azimuth' * band (band) <U3 156B 'b01' 'b02' ... 'b11' 'b12' * detector (detector) int64 40B 8 9 10 11 12 * x (x) int64 184B 300000 305000 ... 410000 * y (y) int64 184B 3100020 3095020 ... 2990020 Data variables: mean_sun_angles (angle) float64 16B dask.array<chunksize=(2,), meta=np.ndarray> mean_viewing_incidence_angles (band, angle) float64 208B dask.array<chunksize=(13, 2), meta=np.ndarray> sun_angles (angle, y, x) float64 8kB dask.array<chunksize=(2, 23, 23), meta=np.ndarray> viewing_incidence_angles (band, detector, angle, y, x) float64 550kB dask.array<chunksize=(7, 5, 2, 23, 23), meta=np.ndarray>geometry- angle: 2
- band: 13
- y: 23
- x: 23
- detector: 5
- angle(angle)<U7'zenith' 'azimuth'
array(['zenith', 'azimuth'], dtype='<U7')
- band(band)<U3'b01' 'b02' 'b03' ... 'b11' 'b12'
array(['b01', 'b02', 'b03', 'b04', 'b05', 'b06', 'b07', 'b08', 'b8a', 'b09', 'b10', 'b11', 'b12'], dtype='<U3') - detector(detector)int648 9 10 11 12
array([ 8, 9, 10, 11, 12])
- x(x)int64300000 305000 ... 405000 410000
array([300000, 305000, 310000, 315000, 320000, 325000, 330000, 335000, 340000, 345000, 350000, 355000, 360000, 365000, 370000, 375000, 380000, 385000, 390000, 395000, 400000, 405000, 410000]) - y(y)int643100020 3095020 ... 2995020 2990020
array([3100020, 3095020, 3090020, 3085020, 3080020, 3075020, 3070020, 3065020, 3060020, 3055020, 3050020, 3045020, 3040020, 3035020, 3030020, 3025020, 3020020, 3015020, 3010020, 3005020, 3000020, 2995020, 2990020])
- mean_sun_angles(angle)float64dask.array<chunksize=(2,), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['angle'], 'dimensions': ['angle']}
- unit :
- deg
Array Chunk Bytes 16 B 16 B Shape (2,) (2,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - mean_viewing_incidence_angles(band, angle)float64dask.array<chunksize=(13, 2), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['angle', 'band'], 'dimensions': ['band', 'angle']}
- unit :
- deg
Array Chunk Bytes 208 B 208 B Shape (13, 2) (13, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - sun_angles(angle, y, x)float64dask.array<chunksize=(2, 23, 23), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['angle', 'y', 'x'], 'dimensions': ['angle', 'y', 'x']}
Array Chunk Bytes 8.27 kiB 8.27 kiB Shape (2, 23, 23) (2, 23, 23) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - viewing_incidence_angles(band, detector, angle, y, x)float64dask.array<chunksize=(7, 5, 2, 23, 23), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['angle', 'y', 'x', 'detector', 'band'], 'dimensions': ['band', 'detector', 'angle', 'y', 'x']}
Array Chunk Bytes 537.27 kiB 289.30 kiB Shape (13, 5, 2, 23, 23) (7, 5, 2, 23, 23) Dask graph 2 chunks in 2 graph layers Data type float64 numpy.ndarray
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*mask<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*detector_footprint<xarray.DatasetView> Size: 482MB Dimensions: (y: 10980, x: 10980) Coordinates: * x (x) int64 88kB 300005 300015 300025 300035 ... 409775 409785 409795 * y (y) int64 88kB 3100015 3100005 3099995 ... 2990245 2990235 2990225 Data variables: b02 (y, x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> b03 (y, x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> b04 (y, x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> b08 (y, x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray>r10m- y: 10980
- x: 10980
- x(x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- y(y)int643100015 3100005 ... 2990235 2990225
array([3100015, 3100005, 3099995, ..., 2990245, 2990235, 2990225])
- b02(y, x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b03(y, x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b04(y, x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b08(y, x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 181MB Dimensions: (y: 5490, x: 5490) Coordinates: * x (x) int64 44kB 300010 300030 300050 300070 ... 409750 409770 409790 * y (y) int64 44kB 3100010 3099990 3099970 ... 2990270 2990250 2990230 Data variables: b05 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b06 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b07 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b11 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b12 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b8a (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray>r20m- y: 5490
- x: 5490
- x(x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- y(y)int643100010 3099990 ... 2990250 2990230
array([3100010, 3099990, 3099970, ..., 2990270, 2990250, 2990230])
- b05(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b06(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b07(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b11(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b12(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b8a(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 10MB Dimensions: (y: 1830, x: 1830) Coordinates: * x (x) int64 15kB 300030 300090 300150 300210 ... 409650 409710 409770 * y (y) int64 15kB 3099990 3099930 3099870 ... 2990370 2990310 2990250 Data variables: b01 (y, x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray> b09 (y, x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray> b10 (y, x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray>r60m- y: 1830
- x: 1830
- x(x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- y(y)int643099990 3099930 ... 2990310 2990250
array([3099990, 3099930, 3099870, ..., 2990370, 2990310, 2990250])
- b01(y, x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b09(y, x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b10(y, x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*l1c_classification<xarray.DatasetView> Size: 3MB Dimensions: (y: 1830, x: 1830) Coordinates: * x (x) int64 15kB 300030 300090 300150 300210 ... 409650 409710 409770 * y (y) int64 15kB 3099990 3099930 3099870 ... 2990370 2990310 2990250 Data variables: b00 (y, x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray>r60m- y: 1830
- x: 1830
- x(x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- y(y)int643099990 3099930 ... 2990310 2990250
array([3099990, 3099930, 3099870, ..., 2990370, 2990310, 2990250])
- b00(y, x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4], 'flag_meanings': ['OPAQUE', 'CIRRUS', 'SNOW_ICE'], 'long_name': 'cloud classification mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4]
- flag_meanings :
- ['OPAQUE', 'CIRRUS', 'SNOW_ICE']
- long_name :
- cloud classification mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*l2a_classification<xarray.DatasetView> Size: 30MB Dimensions: (y: 5490, x: 5490) Coordinates: * x (x) int64 44kB 300010 300030 300050 300070 ... 409750 409770 409790 * y (y) int64 44kB 3100010 3099990 3099970 ... 2990270 2990250 2990230 Data variables: scl (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray>r20m- y: 5490
- x: 5490
- x(x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- y(y)int643100010 3099990 ... 2990250 2990230
array([3100010, 3099990, 3099970, ..., 2990270, 2990250, 2990230])
- scl(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 3MB Dimensions: (y: 1830, x: 1830) Coordinates: * x (x) int64 15kB 300030 300090 300150 300210 ... 409650 409710 409770 * y (y) int64 15kB 3099990 3099930 3099870 ... 2990370 2990310 2990250 Data variables: scl (y, x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray>r60m- y: 1830
- x: 1830
- x(x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- y(y)int643099990 3099930 ... 2990310 2990250
array([3099990, 3099930, 3099870, ..., 2990370, 2990310, 2990250])
- scl(y, x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*meteorology<xarray.DatasetView> Size: 4kB Dimensions: (latitude: 9, longitude: 9) Coordinates: isobaricInhPa float64 8B ... * latitude (latitude) float64 72B 28.01 27.89 27.77 ... 27.16 27.03 * longitude (longitude) float64 72B -11.03 -10.89 ... -10.05 -9.909 number int64 8B ... step int64 8B ... surface float64 8B ... time datetime64[ns] 8B ... valid_time datetime64[ns] 8B ... Data variables: aod1240 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> aod469 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> aod550 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> aod670 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> aod865 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> bcaod550 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> duaod550 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> omaod550 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> ssaod550 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> suaod550 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> z (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> Attributes: (7)cams- latitude: 9
- longitude: 9
- isobaricInhPa()float64...
- long_name :
- pressure
- positive :
- down
- standard_name :
- air_pressure
- stored_direction :
- decreasing
- units :
- hPa
[1 values with dtype=float64]
- latitude(latitude)float6428.01 27.89 27.77 ... 27.16 27.03
- long_name :
- latitude
- standard_name :
- latitude
- stored_direction :
- decreasing
- units :
- degrees_north
array([28.01 , 27.888, 27.766, 27.644, 27.522, 27.4 , 27.278, 27.156, 27.031])
- longitude(longitude)float64-11.03 -10.89 ... -10.05 -9.909
- long_name :
- longitude
- standard_name :
- longitude
- units :
- degrees_east
array([-11.034 , -10.893375, -10.75275 , -10.612125, -10.4715 , -10.330875, -10.19025 , -10.049625, -9.909 ]) - number()int64...
- long_name :
- ensemble member numerical id
- standard_name :
- realization
- units :
- 1
[1 values with dtype=int64]
- step()int64...
- long_name :
- time since forecast_reference_time
- standard_name :
- forecast_period
- units :
- nanoseconds
[1 values with dtype=int64]
- surface()float64...
- long_name :
- original GRIB coordinate for key: level(surface)
- units :
- 1
[1 values with dtype=float64]
- time()datetime64[ns]...
- long_name :
- initial time of forecast
- standard_name :
- forecast_reference_time
[1 values with dtype=datetime64[ns]]
- valid_time()datetime64[ns]...
- long_name :
- time
- standard_name :
- time
[1 values with dtype=datetime64[ns]]
- aod1240(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod1240
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 1240nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210216
- GRIB_shortName :
- aod1240
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 1240nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 1240nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - aod469(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod469
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 469nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210213
- GRIB_shortName :
- aod469
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 469nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 469nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - aod550(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210207
- GRIB_shortName :
- aod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - aod670(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod670
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 670nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210214
- GRIB_shortName :
- aod670
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 670nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 670nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - aod865(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod865
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 865nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210215
- GRIB_shortName :
- aod865
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 865nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 865nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - bcaod550(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- bcaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Black Carbon Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210211
- GRIB_shortName :
- bcaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Black Carbon Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Black Carbon Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - duaod550(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- duaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Dust Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210209
- GRIB_shortName :
- duaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- isobaricInhPa
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Dust Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Dust Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - omaod550(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- omaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Organic Matter Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210210
- GRIB_shortName :
- omaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Organic Matter Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Organic Matter Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - ssaod550(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- ssaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Sea Salt Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210208
- GRIB_shortName :
- ssaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Sea Salt Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Sea Salt Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - suaod550(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- suaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Sulphate Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210212
- GRIB_shortName :
- suaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Sulphate Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Sulphate Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - z(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- geopotential
- GRIB_cfVarName :
- z
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Geopotential
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 129
- GRIB_shortName :
- z
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- m**2 s**-2
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Geopotential', 'standard_name': 'geopotential', 'units': 'm**2 s**-2'}
- long_name :
- Geopotential
- standard_name :
- geopotential
- units :
- m**2 s**-2
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray
- Conventions :
- CF-1.7
- GRIB_centre :
- ecmf
- GRIB_centreDescription :
- European Centre for Medium-Range Weather Forecasts
- GRIB_edition :
- 1
- GRIB_subCentre :
- 0
- history :
- 2025-05-18T20:19 GRIB to CDM+CF via cfgrib-0.9.10.4/ecCodes-2.34.1 with {"source": "../../mnt/data/eopf-conversion-kwr8p/S2B_MSIL2A_20250518T112119_N0511_R037_T29RLL_20250518T140519.SAFE/GRANULE/L2A_T29RLL_A042820_20250518T113246/AUX_DATA/AUX_CAMSFO", "filter_by_keys": {}, "encode_cf": ["parameter", "time", "geography", "vertical"]}
- institution :
- European Centre for Medium-Range Weather Forecasts
<xarray.DatasetView> Size: 2kB Dimensions: (latitude: 9, longitude: 9) Coordinates: isobaricInhPa float64 8B ... * latitude (latitude) float64 72B 28.01 27.89 27.77 ... 27.16 27.03 * longitude (longitude) float64 72B -11.03 -10.89 ... -10.05 -9.909 number int64 8B ... step int64 8B ... surface float64 8B ... time datetime64[ns] 8B ... valid_time datetime64[ns] 8B ... Data variables: msl (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> r (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> tco3 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> tcwv (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> u10 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> v10 (latitude, longitude) float32 324B dask.array<chunksize=(9, 9), meta=np.ndarray> Attributes: (7)ecmwf- latitude: 9
- longitude: 9
- isobaricInhPa()float64...
- long_name :
- pressure
- positive :
- down
- standard_name :
- air_pressure
- stored_direction :
- decreasing
- units :
- hPa
[1 values with dtype=float64]
- latitude(latitude)float6428.01 27.89 27.77 ... 27.16 27.03
- long_name :
- latitude
- standard_name :
- latitude
- stored_direction :
- decreasing
- units :
- degrees_north
array([28.01 , 27.888, 27.766, 27.644, 27.522, 27.4 , 27.278, 27.156, 27.031])
- longitude(longitude)float64-11.03 -10.89 ... -10.05 -9.909
- long_name :
- longitude
- standard_name :
- longitude
- units :
- degrees_east
array([-11.034 , -10.893375, -10.75275 , -10.612125, -10.4715 , -10.330875, -10.19025 , -10.049625, -9.909 ]) - number()int64...
- long_name :
- ensemble member numerical id
- standard_name :
- realization
- units :
- 1
[1 values with dtype=int64]
- step()int64...
- long_name :
- time since forecast_reference_time
- standard_name :
- forecast_period
- units :
- nanoseconds
[1 values with dtype=int64]
- surface()float64...
- long_name :
- original GRIB coordinate for key: level(surface)
- units :
- 1
[1 values with dtype=float64]
- time()datetime64[ns]...
- long_name :
- initial time of forecast
- standard_name :
- forecast_reference_time
[1 values with dtype=datetime64[ns]]
- valid_time()datetime64[ns]...
- long_name :
- time
- standard_name :
- time
[1 values with dtype=datetime64[ns]]
- msl(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- air_pressure_at_mean_sea_level
- GRIB_cfVarName :
- msl
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Mean sea level pressure
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 151
- GRIB_shortName :
- msl
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- Pa
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Mean sea level pressure', 'standard_name': 'air_pressure_at_mean_sea_level', 'units': 'Pa'}
- long_name :
- Mean sea level pressure
- standard_name :
- air_pressure_at_mean_sea_level
- units :
- Pa
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - r(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- relative_humidity
- GRIB_cfVarName :
- r
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Relative humidity
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 157
- GRIB_shortName :
- r
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- isobaricInhPa
- GRIB_units :
- %
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Relative humidity', 'standard_name': 'relative_humidity', 'units': '%'}
- long_name :
- Relative humidity
- standard_name :
- relative_humidity
- units :
- %
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - tco3(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- atmosphere_mass_content_of_ozone
- GRIB_cfVarName :
- tco3
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total column ozone
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 206
- GRIB_shortName :
- tco3
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- kg m**-2
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total column ozone', 'standard_name': 'atmosphere_mass_content_of_ozone', 'units': 'kg m**-2'}
- long_name :
- Total column ozone
- standard_name :
- atmosphere_mass_content_of_ozone
- units :
- kg m**-2
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - tcwv(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- lwe_thickness_of_atmosphere_mass_content_of_water_vapor
- GRIB_cfVarName :
- tcwv
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total column vertically-integrated water vapour
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 137
- GRIB_shortName :
- tcwv
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- kg m**-2
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total column vertically-integrated water vapour', 'standard_name': 'lwe_thickness_of_atmosphere_mass_content_of_water_vapor', 'units': 'kg m**-2'}
- long_name :
- Total column vertically-integrated water vapour
- standard_name :
- lwe_thickness_of_atmosphere_mass_content_of_water_vapor
- units :
- kg m**-2
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - u10(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- u10
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- 10 metre U wind component
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 165
- GRIB_shortName :
- 10u
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- m s**-1
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': '10 metre U wind component', 'standard_name': 'unknown', 'units': 'm s**-1'}
- long_name :
- 10 metre U wind component
- standard_name :
- unknown
- units :
- m s**-1
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - v10(latitude, longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- v10
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.141
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.122
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 28.01
- GRIB_latitudeOfLastGridPointInDegrees :
- 27.031
- GRIB_longitudeOfFirstGridPointInDegrees :
- -11.034
- GRIB_longitudeOfLastGridPointInDegrees :
- -9.909
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- 10 metre V wind component
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 166
- GRIB_shortName :
- 10v
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- m s**-1
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': '10 metre V wind component', 'standard_name': 'unknown', 'units': 'm s**-1'}
- long_name :
- 10 metre V wind component
- standard_name :
- unknown
- units :
- m s**-1
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray
- Conventions :
- CF-1.7
- GRIB_centre :
- ecmf
- GRIB_centreDescription :
- European Centre for Medium-Range Weather Forecasts
- GRIB_edition :
- 1
- GRIB_subCentre :
- 0
- history :
- 2025-05-18T20:19 GRIB to CDM+CF via cfgrib-0.9.10.4/ecCodes-2.34.1 with {"source": "../../mnt/data/eopf-conversion-kwr8p/S2B_MSIL2A_20250518T112119_N0511_R037_T29RLL_20250518T140519.SAFE/GRANULE/L2A_T29RLL_A042820_20250518T113246/AUX_DATA/AUX_ECMWFT", "filter_by_keys": {}, "encode_cf": ["parameter", "time", "geography", "vertical"]}
- institution :
- European Centre for Medium-Range Weather Forecasts
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*measurements<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*reflectance<xarray.DatasetView> Size: 4GB Dimensions: (y: 10980, x: 10980) Coordinates: * x (x) int64 88kB 300005 300015 300025 300035 ... 409775 409785 409795 * y (y) int64 88kB 3100015 3100005 3099995 ... 2990245 2990235 2990225 Data variables: b02 (y, x) float64 964MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> b03 (y, x) float64 964MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> b04 (y, x) float64 964MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> b08 (y, x) float64 964MB dask.array<chunksize=(1830, 1830), meta=np.ndarray>r10m- y: 10980
- x: 10980
- x(x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- y(y)int643100015 3100005 ... 2990235 2990225
array([3100015, 3100005, 3099995, ..., 2990245, 2990235, 2990225])
- b02(y, x)float64dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b02 492.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b02 492.3 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 919.80 MiB 25.55 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b03(y, x)float64dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b03 559.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b03 559.0 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 919.80 MiB 25.55 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b04(y, x)float64dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b04 665.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b04 665.0 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 919.80 MiB 25.55 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b08(y, x)float64dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b08 833.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b08 833.0 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 919.80 MiB 25.55 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray
<xarray.DatasetView> Size: 2GB Dimensions: (y: 5490, x: 5490) Coordinates: * x (x) int64 44kB 300010 300030 300050 300070 ... 409750 409770 409790 * y (y) int64 44kB 3100010 3099990 3099970 ... 2990270 2990250 2990230 Data variables: b01 (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray> b02 (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray> b03 (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray> b04 (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray> b05 (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray> b06 (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray> b07 (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray> b11 (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray> b12 (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray> b8a (y, x) float64 241MB dask.array<chunksize=(915, 915), meta=np.ndarray>r20m- y: 5490
- x: 5490
- x(x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- y(y)int643100010 3099990 ... 2990250 2990230
array([3100010, 3099990, 3099970, ..., 2990270, 2990250, 2990230])
- b01(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b01 442.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b01 442.3 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b02(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b02 492.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b02 492.3 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b03(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b03 559.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b03 559.0 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b04(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b04 665.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b04 665.0 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b05(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b05 703.8 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b05 703.8 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b06(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b06 739.1 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b06 739.1 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b07(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b07 779.7 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b07 779.7 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b11(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b11 1610.4 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b11 1610.4 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b12(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b12 2185.7 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b12 2185.7 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b8a(y, x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b8a 864.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b8a 864.0 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray
<xarray.DatasetView> Size: 295MB Dimensions: (y: 1830, x: 1830) Coordinates: * x (x) int64 15kB 300030 300090 300150 300210 ... 409650 409710 409770 * y (y) int64 15kB 3099990 3099930 3099870 ... 2990370 2990310 2990250 Data variables: b01 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b02 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b03 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b04 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b05 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b06 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b07 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b09 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b11 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b12 (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray> b8a (y, x) float64 27MB dask.array<chunksize=(305, 305), meta=np.ndarray>r60m- y: 1830
- x: 1830
- x(x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- y(y)int643099990 3099930 ... 2990310 2990250
array([3099990, 3099930, 3099870, ..., 2990370, 2990310, 2990250])
- b01(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b01 442.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b01 442.3 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b02(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b02 492.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b02 492.3 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b03(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b03 559.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b03 559.0 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b04(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b04 665.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b04 665.0 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b05(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b05 703.8 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b05 703.8 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b06(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b06 739.1 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b06 739.1 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b07(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b07 779.7 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b07 779.7 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b09(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b09 943.2 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b09 943.2 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b11(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b11 1610.4 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b11 1610.4 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b12(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b12 2185.7 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b12 2185.7 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - b8a(y, x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b8a 864.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b8a 864.0 nm
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*quality<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*atmosphere<xarray.DatasetView> Size: 482MB Dimensions: (y: 10980, x: 10980) Coordinates: * x (x) int64 88kB 300005 300015 300025 300035 ... 409775 409785 409795 * y (y) int64 88kB 3100015 3100005 3099995 ... 2990245 2990235 2990225 Data variables: aot (y, x) uint16 241MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> wvp (y, x) uint16 241MB dask.array<chunksize=(1830, 1830), meta=np.ndarray>r10m- y: 10980
- x: 10980
- x(x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- y(y)int643100015 3100005 ... 2990235 2990225
array([3100015, 3100005, 3099995, ..., 2990245, 2990235, 2990225])
- aot(y, x)uint16dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray - wvp(y, x)uint16dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray
<xarray.DatasetView> Size: 121MB Dimensions: (y: 5490, x: 5490) Coordinates: * x (x) int64 44kB 300010 300030 300050 300070 ... 409750 409770 409790 * y (y) int64 44kB 3100010 3099990 3099970 ... 2990270 2990250 2990230 Data variables: aot (y, x) uint16 60MB dask.array<chunksize=(915, 915), meta=np.ndarray> wvp (y, x) uint16 60MB dask.array<chunksize=(915, 915), meta=np.ndarray>r20m- y: 5490
- x: 5490
- x(x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- y(y)int643100010 3099990 ... 2990250 2990230
array([3100010, 3099990, 3099970, ..., 2990270, 2990250, 2990230])
- aot(y, x)uint16dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 57.49 MiB 1.60 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray - wvp(y, x)uint16dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 57.49 MiB 1.60 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray
<xarray.DatasetView> Size: 13MB Dimensions: (y: 1830, x: 1830) Coordinates: * x (x) int64 15kB 300030 300090 300150 300210 ... 409650 409710 409770 * y (y) int64 15kB 3099990 3099930 3099870 ... 2990370 2990310 2990250 Data variables: aot (y, x) uint16 7MB dask.array<chunksize=(305, 305), meta=np.ndarray> wvp (y, x) uint16 7MB dask.array<chunksize=(305, 305), meta=np.ndarray>r60m- y: 1830
- x: 1830
- x(x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- y(y)int643099990 3099930 ... 2990310 2990250
array([3099990, 3099930, 3099870, ..., 2990370, 2990310, 2990250])
- aot(y, x)uint16dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 6.39 MiB 181.69 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray - wvp(y, x)uint16dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 6.39 MiB 181.69 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*l2a_quicklook<xarray.DatasetView> Size: 362MB Dimensions: (band: 3, y: 10980, x: 10980) Coordinates: * band (band) int64 24B 1 2 3 * x (x) int64 88kB 300005 300015 300025 300035 ... 409775 409785 409795 * y (y) int64 88kB 3100015 3100005 3099995 ... 2990245 2990235 2990225 Data variables: tci (band, y, x) uint8 362MB dask.array<chunksize=(1, 1830, 1830), meta=np.ndarray>r10m- band: 3
- y: 10980
- x: 10980
- band(band)int641 2 3
array([1, 2, 3])
- x(x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- y(y)int643100015 3100005 ... 2990235 2990225
array([3100015, 3100005, 3099995, ..., 2990245, 2990235, 2990225])
- tci(band, y, x)uint8dask.array<chunksize=(1, 1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 344.93 MiB 3.19 MiB Shape (3, 10980, 10980) (1, 1830, 1830) Dask graph 108 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 91MB Dimensions: (band: 3, y: 5490, x: 5490) Coordinates: * band (band) int64 24B 1 2 3 * x (x) int64 44kB 300010 300030 300050 300070 ... 409750 409770 409790 * y (y) int64 44kB 3100010 3099990 3099970 ... 2990270 2990250 2990230 Data variables: tci (band, y, x) uint8 90MB dask.array<chunksize=(1, 915, 915), meta=np.ndarray>r20m- band: 3
- y: 5490
- x: 5490
- band(band)int641 2 3
array([1, 2, 3])
- x(x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- y(y)int643100010 3099990 ... 2990250 2990230
array([3100010, 3099990, 3099970, ..., 2990270, 2990250, 2990230])
- tci(band, y, x)uint8dask.array<chunksize=(1, 915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 86.23 MiB 817.60 kiB Shape (3, 5490, 5490) (1, 915, 915) Dask graph 108 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 10MB Dimensions: (band: 3, y: 1830, x: 1830) Coordinates: * band (band) int64 24B 1 2 3 * x (x) int64 15kB 300030 300090 300150 300210 ... 409650 409710 409770 * y (y) int64 15kB 3099990 3099930 3099870 ... 2990370 2990310 2990250 Data variables: tci (band, y, x) uint8 10MB dask.array<chunksize=(1, 305, 305), meta=np.ndarray>r60m- band: 3
- y: 1830
- x: 1830
- band(band)int641 2 3
array([1, 2, 3])
- x(x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- y(y)int643099990 3099930 ... 2990310 2990250
array([3099990, 3099930, 3099870, ..., 2990370, 2990310, 2990250])
- tci(band, y, x)uint8dask.array<chunksize=(1, 305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 9.58 MiB 90.84 kiB Shape (3, 1830, 1830) (1, 305, 305) Dask graph 108 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*mask<xarray.DatasetView> Size: 482MB Dimensions: (y: 10980, x: 10980) Coordinates: * x (x) int64 88kB 300005 300015 300025 300035 ... 409775 409785 409795 * y (y) int64 88kB 3100015 3100005 3099995 ... 2990245 2990235 2990225 Data variables: b02 (y, x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> b03 (y, x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> b04 (y, x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray> b08 (y, x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray>r10m- y: 10980
- x: 10980
- x(x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- y(y)int643100015 3100005 ... 2990235 2990225
array([3100015, 3100005, 3099995, ..., 2990245, 2990235, 2990225])
- b02(y, x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b03(y, x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b04(y, x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b08(y, x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 181MB Dimensions: (y: 5490, x: 5490) Coordinates: * x (x) int64 44kB 300010 300030 300050 300070 ... 409750 409770 409790 * y (y) int64 44kB 3100010 3099990 3099970 ... 2990270 2990250 2990230 Data variables: b05 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b06 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b07 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b11 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b12 (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> b8a (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray>r20m- y: 5490
- x: 5490
- x(x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- y(y)int643100010 3099990 ... 2990250 2990230
array([3100010, 3099990, 3099970, ..., 2990270, 2990250, 2990230])
- b05(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b06(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b07(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b11(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b12(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b8a(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 10MB Dimensions: (y: 1830, x: 1830) Coordinates: * x (x) int64 15kB 300030 300090 300150 300210 ... 409650 409710 409770 * y (y) int64 15kB 3099990 3099930 3099870 ... 2990370 2990310 2990250 Data variables: b01 (y, x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray> b09 (y, x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray> b10 (y, x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray>r60m- y: 1830
- x: 1830
- x(x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- y(y)int643099990 3099930 ... 2990310 2990250
array([3099990, 3099930, 3099870, ..., 2990370, 2990310, 2990250])
- b01(y, x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b09(y, x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - b10(y, x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*probability<xarray.DatasetView> Size: 60MB Dimensions: (y: 5490, x: 5490) Coordinates: band int64 8B ... * x (x) int64 44kB 300010 300030 300050 300070 ... 409750 409770 409790 * y (y) int64 44kB 3100010 3099990 3099970 ... 2990270 2990250 2990230 Data variables: cld (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray> snw (y, x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray>r20m- y: 5490
- x: 5490
- band()int64...
[1 values with dtype=int64]
- x(x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- y(y)int643100010 3099990 ... 2990250 2990230
array([3100010, 3099990, 3099970, ..., 2990270, 2990250, 2990230])
- cld(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - snw(y, x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
- other_metadata :
- {'AOT_retrieval_model': 'CAMS', 'L0_ancillary_data_quality': '2', 'L0_ephemeris_data_quality': '3', 'NUC_table_ID': 2, 'SWIR_rearrangement_flag': 'null', 'UTM_zone_identification': 'S2B_OPER_MSI_L2A_TL_2BPS_20250518T140519_A042820_T29RLL_N05.11', 'absolute_location_assessment_from_AOCS': '\n ', 'band_description': {'01': {'bandwidth': '20.0', 'central_wavelength': 442.3, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '7.22833', 'physical_gain': '3.958028', 'spectral_response_step': '1', 'spectral_response_values': '0.0062411 0.01024045 0.00402983 0.00642179 0.00552753 0.0065525 0.00409887 0.006297 0.00436742 0.00233356 0.00058162 0.00202276 0.00294328 0.00485362 0.00317041 0.00237657 0.00234612 0.00440152 0.01292397 0.05001678 0.18650104 0.45441623 0.72307877 0.83999211 0.86456334 0.87472096 0.89215296 0.91090814 0.92588017 0.93924094 0.94491826 0.95078529 0.96803023 0.99939195 1 0.97548364 0.96148351 0.94986211 0.91841452 0.87989802 0.80383677 0.59752075 0.30474132 0.10798014 0.0304465 0.00885119', 'units': 'nm', 'wavelength_max': 456.0, 'wavelength_min': 411.0}, '02': {'bandwidth': '65.0', 'central_wavelength': 492.3, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.2646269', 'physical_gain': '3.81509366', 'spectral_response_step': '1', 'spectral_response_values': '0.05529541 0.12005068 0.25199051 0.4623617 0.65162379 0.77642171 0.82319091 0.83083116 0.83382106 0.837526 0.86304286 0.88226141 0.90486326 0.92043837 0.93602675 0.930533 0.92714067 0.9161479 0.90551724 0.89745515 0.90266694 0.90854264 0.92047913 0.92417935 0.91845025 0.90743244 0.89733983 0.88646415 0.87189983 0.85643973 0.84473414 0.84190734 0.85644111 0.87782724 0.90261174 0.91840544 0.94585847 0.96887192 0.99336135 0.99927899 1 0.99520325 0.98412711 0.97947473 0.97808297 0.97213439 0.96277794 0.95342234 0.93802376 0.92460144 0.90932642 0.90192251 0.89184298 0.88963556 0.89146958 0.89877911 0.91056869 0.92427362 0.93823555 0.95311791 0.97150808 0.98737003 0.99658514 0.99367959 0.98144714 0.95874415 0.89291635 0.73566218 0.52060373 0.3322804 0.19492197 0.11732617 0.07507304 0.05094154 0.03213016 0.01510217 0.00447984', 'units': 'nm', 'wavelength_max': 532.0, 'wavelength_min': 456.0}, '03': {'bandwidth': '35.0', 'central_wavelength': 559.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.2996743', 'physical_gain': '4.23219561', 'spectral_response_step': '1', 'spectral_response_values': '0.00188039 0.01545903 0.03660414 0.08100583 0.16917887 0.33278274 0.58622794 0.80916412 0.913051 0.94472284 0.94898813 0.94369132 0.92845674 0.91256938 0.90078036 0.89958598 0.90547138 0.92045355 0.94065665 0.96199681 0.98186744 0.9985841 1 0.99279888 0.97801325 0.95301174 0.9266333 0.89359131 0.86941793 0.84827 0.83908301 0.83206209 0.8291787 0.83305844 0.84630939 0.86396307 0.87268076 0.86818339 0.8554947 0.80839054 0.67650876 0.45584205 0.24737576 0.12765465 0.0589016 0.02564742 0.00515905', 'units': 'nm', 'wavelength_max': 582.0, 'wavelength_min': 536.0}, '04': {'bandwidth': '30.0', 'central_wavelength': 665.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.3405629', 'physical_gain': '4.77028146', 'spectral_response_step': '1', 'spectral_response_values': '0.00499358 0.02642563 0.11905127 0.333204 0.59813448 0.80612041 0.91152955 0.92179127 0.91677167 0.90751672 0.89867522 0.89413622 0.89685141 0.89933396 0.90191681 0.90710817 0.9164622 0.92908702 0.9426682 0.95591935 0.96854537 0.98264967 0.99231022 1 0.99904114 0.99257339 0.97943242 0.96553214 0.95377013 0.94146127 0.92151286 0.89308475 0.83539461 0.69759082 0.49483622 0.27886075 0.10892715 0.03028701 0.00747382 0.00087148', 'units': 'nm', 'wavelength_max': 685.0, 'wavelength_min': 646.0}, '05': {'bandwidth': '15.0', 'central_wavelength': 703.8, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7846026', 'physical_gain': '5.17738824', 'spectral_response_step': '1', 'spectral_response_values': '0.01042619 0.05713826 0.21461286 0.54715702 0.87088164 0.96808183 0.99104427 1 0.99512875 0.98751456 0.97910038 0.97035979 0.95875454 0.94130694 0.92531149 0.89283152 0.76531084 0.50228771 0.17957688 0.0337948 0.00240526', 'units': 'nm', 'wavelength_max': 714.0, 'wavelength_min': 694.0}, '06': {'bandwidth': '13.0', 'central_wavelength': 739.1, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7727115', 'physical_gain': '5.08794688', 'spectral_response_step': '1', 'spectral_response_values': '0.01739744 0.10565746 0.38571484 0.78168196 0.90518378 0.91562509 0.92258804 0.93134141 0.9469604 0.96535098 0.97817455 0.99107716 0.99990615 1 0.97144118 0.81937503 0.46748011 0.09409351 0.00983236', 'units': 'nm', 'wavelength_max': 748.0, 'wavelength_min': 730.0}, '07': {'bandwidth': '19.0', 'central_wavelength': 779.7, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7727115', 'physical_gain': '4.76255482', 'spectral_response_step': '1', 'spectral_response_values': '0.0103729 0.03754921 0.11240409 0.25890105 0.48035521 0.73155954 0.91293607 0.97124929 0.96391197 0.95529249 0.964831 0.98628988 1 0.99782157 0.98343012 0.96489467 0.94619093 0.92560158 0.90788186 0.88471259 0.85693094 0.82513165 0.7734046 0.66767522 0.47756609 0.23225321 0.06764032 0.01301456 0.00117425', 'units': 'nm', 'wavelength_max': 794.0, 'wavelength_min': 766.0}, '08': {'bandwidth': '105.0', 'central_wavelength': 833.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.3872929', 'physical_gain': '6.82093912', 'spectral_response_step': '1', 'spectral_response_values': '0.00037316 0.00296451 0.01663315 0.02818619 0.05000442 0.08575595 0.14871265 0.25701156 0.42307501 0.62943997 0.80112571 0.90352196 0.93855197 0.94406104 0.9474892 0.96325767 0.97948137 0.99144397 0.99630748 1 0.99827121 0.99843182 0.98914342 0.98264167 0.96769944 0.95752283 0.95074919 0.9458125 0.94267916 0.9465958 0.94450012 0.93992861 0.92759688 0.91226544 0.89079677 0.8706102 0.85021777 0.83416655 0.82214927 0.8124078 0.80920229 0.80220847 0.79081499 0.78239761 0.76731527 0.75394962 0.74226922 0.72750987 0.71976209 0.71456726 0.71982866 0.72746214 0.73945306 0.75138424 0.76310661 0.77122498 0.78298312 0.78494127 0.78409222 0.7834498 0.78216032 0.78062401 0.78132572 0.7813272 0.7810081 0.77897938 0.7761445 0.76910534 0.7625494 0.75157186 0.74086146 0.73121299 0.71988688 0.71025573 0.69679744 0.68602501 0.67163906 0.65532408 0.64173681 0.62683353 0.61241074 0.60185411 0.59380689 0.58714687 0.58444579 0.58231388 0.58111599 0.57996902 0.57480451 0.57684802 0.57273034 0.57144461 0.56985127 0.57167225 0.57154913 0.57292235 0.57617796 0.5784908 0.58023702 0.57982619 0.57868642 0.57587451 0.56976789 0.56173136 0.55644176 0.54881732 0.54508423 0.54153848 0.54069902 0.53850959 0.53655263 0.530404 0.52068821 0.50399678 0.486513 0.46813829 0.45468861 0.44447936 0.44177056 0.44425396 0.44633078 0.43914074 0.41748156 0.3690277 0.30165803 0.23504284 0.17434599 0.12247894 0.08354059 0.05624109 0.03804368 0.02427229 0.01490577 0.00615862', 'units': 'nm', 'wavelength_max': 907.0, 'wavelength_min': 774.0}, '09': {'bandwidth': '20.0', 'central_wavelength': 943.2, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '8.031501', 'physical_gain': '9.37557056', 'spectral_response_step': '1', 'spectral_response_values': '0.0121336 0.04608767 0.15156613 0.35888361 0.60704101 0.83836043 0.93474094 0.94270146 0.95838078 0.99064712 0.99789825 1 0.98593726 0.97333604 0.95776631 0.972226 0.94856942 0.94367414 0.90771555 0.88460732 0.85258329 0.83375172 0.71599386 0.52202762 0.26922852 0.09477806 0.02640828 0.00346547', 'units': 'nm', 'wavelength_max': 957.0, 'wavelength_min': 930.0}, '10': {'bandwidth': '30.0', 'central_wavelength': 1376.9, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '5.5537686', 'physical_gain': '56.48916794', 'spectral_response_step': '1', 'spectral_response_values': '2.472e-05 0.00013691 0.00012558 8.901e-05 0.00012425 9.941e-05 0.00013952 0.00015816 0.00019272 0.00025959 0.00032221 0.00034719 0.0003699 0.00054874 0.00105434 0.00218813 0.00480743 0.01135252 0.02671185 0.05776022 0.11176337 0.19587518 0.31418191 0.46188068 0.62292578 0.7709851 0.88086652 0.9448941 0.97405066 0.98616696 0.99306955 0.99775441 1 0.99942348 0.99616891 0.99082045 0.9842131 0.97708513 0.97013647 0.96374366 0.95755001 0.95127438 0.94546638 0.94069659 0.93759595 0.93624612 0.93510206 0.93054472 0.91630845 0.88530334 0.83129653 0.74856466 0.63524397 0.49733159 0.34907723 0.21259735 0.10971453 0.04789269 0.01853013 0.00716776 0.0031533 0.00157017 0.00084901 0.00053006 0.00033171 0.00019447 0.00022104 0.00022646 0.00018156 0.00016063 0.00015475 0.00014734 0.00014776 0.00017405 0.00023619 0.00012007 4.337e-05', 'units': 'nm', 'wavelength_max': 1415.0, 'wavelength_min': 1339.0}, '11': {'bandwidth': '90.0', 'central_wavelength': 1610.4, 'onboard_compression_rate': '2.4', 'onboard_integration_time': '1.3793689', 'physical_gain': '37.20623728', 'spectral_response_step': '1', 'spectral_response_values': '1.154e-05 2.707e-05 8.129e-05 0.0001701 0.00027422 0.00034456 0.00046028 0.00065214 0.00082283 0.00107791 0.0014306 0.00196134 0.00266427 0.00368682 0.00522456 0.00758401 0.01126335 0.01715812 0.02674581 0.04145595 0.06300627 0.09464207 0.13995799 0.20105412 0.28189591 0.38134665 0.4907345 0.60674263 0.71505301 0.80391496 0.87015099 0.91645643 0.94668952 0.96391534 0.97305962 0.97704089 0.97777566 0.97686717 0.97531356 0.97336816 0.9714563 0.9697157 0.96907419 0.96968255 0.97051178 0.97272986 0.97613656 0.97894419 0.9810083 0.98350836 0.9848292 0.98438948 0.98389859 0.98334634 0.9814301 0.97936035 0.97802641 0.97623515 0.97537114 0.97569131 0.97679261 0.97898052 0.98199689 0.98520852 0.98866135 0.99233425 0.99480248 0.99589079 0.9958911 0.99475534 0.99207775 0.98856394 0.9848769 0.98106836 0.97677436 0.97351815 0.97192459 0.97052192 0.97043004 0.9723835 0.97525347 0.97856769 0.98298866 0.98810437 0.99268138 0.99645012 0.9990686 1 0.99902738 0.99662493 0.99326995 0.989647 0.98577051 0.98212932 0.97979728 0.97946062 0.980262 0.98247241 0.98601349 0.98957829 0.99173488 0.99219848 0.98937107 0.98205611 0.97007817 0.95307506 0.93223131 0.90784439 0.88392149 0.86411672 0.85075738 0.84410342 0.84337963 0.84698191 0.84866039 0.84046041 0.81336359 0.75654857 0.66994259 0.56517119 0.45690882 0.35310835 0.25633426 0.17582806 0.11552613 0.0733101 0.04640345 0.02898639 0.01853597 0.01243537 0.00877131 0.00630418 0.00457459 0.00335323 0.00245906 0.001988 0.00149989 0.00112208 0.00078208 0.00054086 0.00028019 0.0001326', 'units': 'nm', 'wavelength_max': 1679.0, 'wavelength_min': 1538.0}, '12': {'bandwidth': '180.0', 'central_wavelength': 2185.7, 'onboard_compression_rate': '2.4', 'onboard_integration_time': '1.4761667', 'physical_gain': '108.65081694', 'spectral_response_step': '1', 'spectral_response_values': '0.00022389 0.00073676 0.00164703 0.00301151 0.00458328 0.00592584 0.00752876 0.00874103 0.01025764 0.01222618 0.01458055 0.01744267 0.02104287 0.02540339 0.03057901 0.03719619 0.04572365 0.05630242 0.06994211 0.08791078 0.11057655 0.13873936 0.17311239 0.21416774 0.26175285 0.31696031 0.38057337 0.44916129 0.52246923 0.59858476 0.67183039 0.73762307 0.79267856 0.83543144 0.86612544 0.88613168 0.89739036 0.90131058 0.90056883 0.89688046 0.89211284 0.88771935 0.88417799 0.88183136 0.88142338 0.88242075 0.88687585 0.89245189 0.89861914 0.90533051 0.91254666 0.91988456 0.92662076 0.93280462 0.9381479 0.94308713 0.94528987 0.94711578 0.94827846 0.94854335 0.94782599 0.94683272 0.94584552 0.94458923 0.94320642 0.94161778 0.94216937 0.94300085 0.94396863 0.94500784 0.94593652 0.94680905 0.94766521 0.94866638 0.94960932 0.95040536 0.95078607 0.95126357 0.95205865 0.9524548 0.95228734 0.95215614 0.95239704 0.95270563 0.95306455 0.9535262 0.95404061 0.9545903 0.95529443 0.95650666 0.95774374 0.95899449 0.96021128 0.96117558 0.96241242 0.96389292 0.9633817 0.96287807 0.96252982 0.96163134 0.96051578 0.95934879 0.95816596 0.95676127 0.95491383 0.95287555 0.95386662 0.95498503 0.9555239 0.95548083 0.9551674 0.95442758 0.95327341 0.95194429 0.95041866 0.9484181 0.94839027 0.94838056 0.94813039 0.94753901 0.94645196 0.94504703 0.9433144 0.94197531 0.94049427 0.93875727 0.94132922 0.9436129 0.94558364 0.94711284 0.94831802 0.94945878 0.95060227 0.95182631 0.95345901 0.95510105 0.95225316 0.95033203 0.94956701 0.94895037 0.94955832 0.95152282 0.95486528 0.95963437 0.96607045 0.97375116 0.97078626 0.96900558 0.96819786 0.96750837 0.9674355 0.96792378 0.96839035 0.96883692 0.9693902 0.96985301 0.96898817 0.96814874 0.96743874 0.96638941 0.96534457 0.96425351 0.96281969 0.96155971 0.96053251 0.95926107 0.95959913 0.96025143 0.96154033 0.96262988 0.96359875 0.96467154 0.9654114 0.96625109 0.96744643 0.96878244 0.97046916 0.97234778 0.97422228 0.9759326 0.97713045 0.97892333 0.98039008 0.98147316 0.98248415 0.9832885 0.98458694 0.9866985 0.98911057 0.99119702 0.99315819 0.99524701 0.99668121 0.99826512 0.99959594 1 0.99866374 0.99576531 0.99062502 0.98155399 0.96876193 0.95174168 0.92942389 0.90240499 0.8705554 0.83207693 0.78626172 0.73649625 0.68202115 0.62329799 0.56418429 0.50680538 0.4515209 0.4006232 0.35430248 0.30995212 0.26941268 0.23418861 0.20232924 0.17373524 0.14941215 0.1290062 0.11157271 0.09693492 0.08490613 0.07409041 0.06449244 0.056343 0.0493999 0.04321207 0.03800099 0.03348242 0.02900748 0.0251013 0.02003645 0.01384563 0.00850471 0.00443484 0.00085324', 'units': 'nm', 'wavelength_max': 2303.0, 'wavelength_min': 2065.0}, '8A': {'bandwidth': '20.0', 'central_wavelength': 864.0, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7608204', 'physical_gain': '5.76107136', 'spectral_response_step': '1', 'spectral_response_values': '0.00167523 0.01602231 0.03225867 0.07345268 0.1689243 0.34543042 0.56923369 0.79611745 0.93749188 0.98102805 0.98742384 0.99457226 0.99912415 0.99993652 1 0.99437257 0.98756135 0.98263615 0.9790323 0.97397518 0.97130259 0.9645338 0.95610202 0.93941552 0.89155652 0.77601041 0.5951886 0.37588812 0.18394037 0.07870072 0.0332686 0.01575167 0.00159818', 'units': 'nm', 'wavelength_max': 880.0, 'wavelength_min': 848.0}, 'b01': {'bandwidth': 20.0, 'central_wavelength': 442.3, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '7.22833', 'physical_gain': '3.958028', 'spectral_response_step': '1', 'spectral_response_values': '0.0062411 0.01024045 0.00402983 0.00642179 0.00552753 0.0065525 0.00409887 0.006297 0.00436742 0.00233356 0.00058162 0.00202276 0.00294328 0.00485362 0.00317041 0.00237657 0.00234612 0.00440152 0.01292397 0.05001678 0.18650104 0.45441623 0.72307877 0.83999211 0.86456334 0.87472096 0.89215296 0.91090814 0.92588017 0.93924094 0.94491826 0.95078529 0.96803023 0.99939195 1 0.97548364 0.96148351 0.94986211 0.91841452 0.87989802 0.80383677 0.59752075 0.30474132 0.10798014 0.0304465 0.00885119', 'units': 'nm', 'wavelength_max': 456.0, 'wavelength_min': 411.0}, 'b02': {'bandwidth': 65.0, 'central_wavelength': 492.3, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.2646269', 'physical_gain': '3.81509366', 'spectral_response_step': '1', 'spectral_response_values': '0.05529541 0.12005068 0.25199051 0.4623617 0.65162379 0.77642171 0.82319091 0.83083116 0.83382106 0.837526 0.86304286 0.88226141 0.90486326 0.92043837 0.93602675 0.930533 0.92714067 0.9161479 0.90551724 0.89745515 0.90266694 0.90854264 0.92047913 0.92417935 0.91845025 0.90743244 0.89733983 0.88646415 0.87189983 0.85643973 0.84473414 0.84190734 0.85644111 0.87782724 0.90261174 0.91840544 0.94585847 0.96887192 0.99336135 0.99927899 1 0.99520325 0.98412711 0.97947473 0.97808297 0.97213439 0.96277794 0.95342234 0.93802376 0.92460144 0.90932642 0.90192251 0.89184298 0.88963556 0.89146958 0.89877911 0.91056869 0.92427362 0.93823555 0.95311791 0.97150808 0.98737003 0.99658514 0.99367959 0.98144714 0.95874415 0.89291635 0.73566218 0.52060373 0.3322804 0.19492197 0.11732617 0.07507304 0.05094154 0.03213016 0.01510217 0.00447984', 'units': 'nm', 'wavelength_max': 532.0, 'wavelength_min': 456.0}, 'b03': {'bandwidth': 35.0, 'central_wavelength': 559.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.2996743', 'physical_gain': '4.23219561', 'spectral_response_step': '1', 'spectral_response_values': '0.00188039 0.01545903 0.03660414 0.08100583 0.16917887 0.33278274 0.58622794 0.80916412 0.913051 0.94472284 0.94898813 0.94369132 0.92845674 0.91256938 0.90078036 0.89958598 0.90547138 0.92045355 0.94065665 0.96199681 0.98186744 0.9985841 1 0.99279888 0.97801325 0.95301174 0.9266333 0.89359131 0.86941793 0.84827 0.83908301 0.83206209 0.8291787 0.83305844 0.84630939 0.86396307 0.87268076 0.86818339 0.8554947 0.80839054 0.67650876 0.45584205 0.24737576 0.12765465 0.0589016 0.02564742 0.00515905', 'units': 'nm', 'wavelength_max': 582.0, 'wavelength_min': 536.0}, 'b04': {'bandwidth': 30.0, 'central_wavelength': 665.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.3405629', 'physical_gain': '4.77028146', 'spectral_response_step': '1', 'spectral_response_values': '0.00499358 0.02642563 0.11905127 0.333204 0.59813448 0.80612041 0.91152955 0.92179127 0.91677167 0.90751672 0.89867522 0.89413622 0.89685141 0.89933396 0.90191681 0.90710817 0.9164622 0.92908702 0.9426682 0.95591935 0.96854537 0.98264967 0.99231022 1 0.99904114 0.99257339 0.97943242 0.96553214 0.95377013 0.94146127 0.92151286 0.89308475 0.83539461 0.69759082 0.49483622 0.27886075 0.10892715 0.03028701 0.00747382 0.00087148', 'units': 'nm', 'wavelength_max': 685.0, 'wavelength_min': 646.0}, 'b05': {'bandwidth': 15.0, 'central_wavelength': 703.8, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7846026', 'physical_gain': '5.17738824', 'spectral_response_step': '1', 'spectral_response_values': '0.01042619 0.05713826 0.21461286 0.54715702 0.87088164 0.96808183 0.99104427 1 0.99512875 0.98751456 0.97910038 0.97035979 0.95875454 0.94130694 0.92531149 0.89283152 0.76531084 0.50228771 0.17957688 0.0337948 0.00240526', 'units': 'nm', 'wavelength_max': 714.0, 'wavelength_min': 694.0}, 'b06': {'bandwidth': 13.0, 'central_wavelength': 739.1, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7727115', 'physical_gain': '5.08794688', 'spectral_response_step': '1', 'spectral_response_values': '0.01739744 0.10565746 0.38571484 0.78168196 0.90518378 0.91562509 0.92258804 0.93134141 0.9469604 0.96535098 0.97817455 0.99107716 0.99990615 1 0.97144118 0.81937503 0.46748011 0.09409351 0.00983236', 'units': 'nm', 'wavelength_max': 748.0, 'wavelength_min': 730.0}, 'b07': {'bandwidth': 19.0, 'central_wavelength': 779.7, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7727115', 'physical_gain': '4.76255482', 'spectral_response_step': '1', 'spectral_response_values': '0.0103729 0.03754921 0.11240409 0.25890105 0.48035521 0.73155954 0.91293607 0.97124929 0.96391197 0.95529249 0.964831 0.98628988 1 0.99782157 0.98343012 0.96489467 0.94619093 0.92560158 0.90788186 0.88471259 0.85693094 0.82513165 0.7734046 0.66767522 0.47756609 0.23225321 0.06764032 0.01301456 0.00117425', 'units': 'nm', 'wavelength_max': 794.0, 'wavelength_min': 766.0}, 'b08': {'bandwidth': 105.0, 'central_wavelength': 833.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.3872929', 'physical_gain': '6.82093912', 'spectral_response_step': '1', 'spectral_response_values': '0.00037316 0.00296451 0.01663315 0.02818619 0.05000442 0.08575595 0.14871265 0.25701156 0.42307501 0.62943997 0.80112571 0.90352196 0.93855197 0.94406104 0.9474892 0.96325767 0.97948137 0.99144397 0.99630748 1 0.99827121 0.99843182 0.98914342 0.98264167 0.96769944 0.95752283 0.95074919 0.9458125 0.94267916 0.9465958 0.94450012 0.93992861 0.92759688 0.91226544 0.89079677 0.8706102 0.85021777 0.83416655 0.82214927 0.8124078 0.80920229 0.80220847 0.79081499 0.78239761 0.76731527 0.75394962 0.74226922 0.72750987 0.71976209 0.71456726 0.71982866 0.72746214 0.73945306 0.75138424 0.76310661 0.77122498 0.78298312 0.78494127 0.78409222 0.7834498 0.78216032 0.78062401 0.78132572 0.7813272 0.7810081 0.77897938 0.7761445 0.76910534 0.7625494 0.75157186 0.74086146 0.73121299 0.71988688 0.71025573 0.69679744 0.68602501 0.67163906 0.65532408 0.64173681 0.62683353 0.61241074 0.60185411 0.59380689 0.58714687 0.58444579 0.58231388 0.58111599 0.57996902 0.57480451 0.57684802 0.57273034 0.57144461 0.56985127 0.57167225 0.57154913 0.57292235 0.57617796 0.5784908 0.58023702 0.57982619 0.57868642 0.57587451 0.56976789 0.56173136 0.55644176 0.54881732 0.54508423 0.54153848 0.54069902 0.53850959 0.53655263 0.530404 0.52068821 0.50399678 0.486513 0.46813829 0.45468861 0.44447936 0.44177056 0.44425396 0.44633078 0.43914074 0.41748156 0.3690277 0.30165803 0.23504284 0.17434599 0.12247894 0.08354059 0.05624109 0.03804368 0.02427229 0.01490577 0.00615862', 'units': 'nm', 'wavelength_max': 907.0, 'wavelength_min': 774.0}, 'b09': {'bandwidth': 20.0, 'central_wavelength': 943.2, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '8.031501', 'physical_gain': '9.37557056', 'spectral_response_step': '1', 'spectral_response_values': '0.0121336 0.04608767 0.15156613 0.35888361 0.60704101 0.83836043 0.93474094 0.94270146 0.95838078 0.99064712 0.99789825 1 0.98593726 0.97333604 0.95776631 0.972226 0.94856942 0.94367414 0.90771555 0.88460732 0.85258329 0.83375172 0.71599386 0.52202762 0.26922852 0.09477806 0.02640828 0.00346547', 'units': 'nm', 'wavelength_max': 957.0, 'wavelength_min': 930.0}, 'b10': {'bandwidth': 30.0, 'central_wavelength': 1376.9, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '5.5537686', 'physical_gain': '56.48916794', 'spectral_response_step': '1', 'spectral_response_values': '2.472e-05 0.00013691 0.00012558 8.901e-05 0.00012425 9.941e-05 0.00013952 0.00015816 0.00019272 0.00025959 0.00032221 0.00034719 0.0003699 0.00054874 0.00105434 0.00218813 0.00480743 0.01135252 0.02671185 0.05776022 0.11176337 0.19587518 0.31418191 0.46188068 0.62292578 0.7709851 0.88086652 0.9448941 0.97405066 0.98616696 0.99306955 0.99775441 1 0.99942348 0.99616891 0.99082045 0.9842131 0.97708513 0.97013647 0.96374366 0.95755001 0.95127438 0.94546638 0.94069659 0.93759595 0.93624612 0.93510206 0.93054472 0.91630845 0.88530334 0.83129653 0.74856466 0.63524397 0.49733159 0.34907723 0.21259735 0.10971453 0.04789269 0.01853013 0.00716776 0.0031533 0.00157017 0.00084901 0.00053006 0.00033171 0.00019447 0.00022104 0.00022646 0.00018156 0.00016063 0.00015475 0.00014734 0.00014776 0.00017405 0.00023619 0.00012007 4.337e-05', 'units': 'nm', 'wavelength_max': 1415.0, 'wavelength_min': 1339.0}, 'b11': {'bandwidth': 90.0, 'central_wavelength': 1610.4, 'onboard_compression_rate': '2.4', 'onboard_integration_time': '1.3793689', 'physical_gain': '37.20623728', 'spectral_response_step': '1', 'spectral_response_values': '1.154e-05 2.707e-05 8.129e-05 0.0001701 0.00027422 0.00034456 0.00046028 0.00065214 0.00082283 0.00107791 0.0014306 0.00196134 0.00266427 0.00368682 0.00522456 0.00758401 0.01126335 0.01715812 0.02674581 0.04145595 0.06300627 0.09464207 0.13995799 0.20105412 0.28189591 0.38134665 0.4907345 0.60674263 0.71505301 0.80391496 0.87015099 0.91645643 0.94668952 0.96391534 0.97305962 0.97704089 0.97777566 0.97686717 0.97531356 0.97336816 0.9714563 0.9697157 0.96907419 0.96968255 0.97051178 0.97272986 0.97613656 0.97894419 0.9810083 0.98350836 0.9848292 0.98438948 0.98389859 0.98334634 0.9814301 0.97936035 0.97802641 0.97623515 0.97537114 0.97569131 0.97679261 0.97898052 0.98199689 0.98520852 0.98866135 0.99233425 0.99480248 0.99589079 0.9958911 0.99475534 0.99207775 0.98856394 0.9848769 0.98106836 0.97677436 0.97351815 0.97192459 0.97052192 0.97043004 0.9723835 0.97525347 0.97856769 0.98298866 0.98810437 0.99268138 0.99645012 0.9990686 1 0.99902738 0.99662493 0.99326995 0.989647 0.98577051 0.98212932 0.97979728 0.97946062 0.980262 0.98247241 0.98601349 0.98957829 0.99173488 0.99219848 0.98937107 0.98205611 0.97007817 0.95307506 0.93223131 0.90784439 0.88392149 0.86411672 0.85075738 0.84410342 0.84337963 0.84698191 0.84866039 0.84046041 0.81336359 0.75654857 0.66994259 0.56517119 0.45690882 0.35310835 0.25633426 0.17582806 0.11552613 0.0733101 0.04640345 0.02898639 0.01853597 0.01243537 0.00877131 0.00630418 0.00457459 0.00335323 0.00245906 0.001988 0.00149989 0.00112208 0.00078208 0.00054086 0.00028019 0.0001326', 'units': 'nm', 'wavelength_max': 1679.0, 'wavelength_min': 1538.0}, 'b12': {'bandwidth': 180.0, 'central_wavelength': 2185.7, 'onboard_compression_rate': '2.4', 'onboard_integration_time': '1.4761667', 'physical_gain': '108.65081694', 'spectral_response_step': '1', 'spectral_response_values': '0.00022389 0.00073676 0.00164703 0.00301151 0.00458328 0.00592584 0.00752876 0.00874103 0.01025764 0.01222618 0.01458055 0.01744267 0.02104287 0.02540339 0.03057901 0.03719619 0.04572365 0.05630242 0.06994211 0.08791078 0.11057655 0.13873936 0.17311239 0.21416774 0.26175285 0.31696031 0.38057337 0.44916129 0.52246923 0.59858476 0.67183039 0.73762307 0.79267856 0.83543144 0.86612544 0.88613168 0.89739036 0.90131058 0.90056883 0.89688046 0.89211284 0.88771935 0.88417799 0.88183136 0.88142338 0.88242075 0.88687585 0.89245189 0.89861914 0.90533051 0.91254666 0.91988456 0.92662076 0.93280462 0.9381479 0.94308713 0.94528987 0.94711578 0.94827846 0.94854335 0.94782599 0.94683272 0.94584552 0.94458923 0.94320642 0.94161778 0.94216937 0.94300085 0.94396863 0.94500784 0.94593652 0.94680905 0.94766521 0.94866638 0.94960932 0.95040536 0.95078607 0.95126357 0.95205865 0.9524548 0.95228734 0.95215614 0.95239704 0.95270563 0.95306455 0.9535262 0.95404061 0.9545903 0.95529443 0.95650666 0.95774374 0.95899449 0.96021128 0.96117558 0.96241242 0.96389292 0.9633817 0.96287807 0.96252982 0.96163134 0.96051578 0.95934879 0.95816596 0.95676127 0.95491383 0.95287555 0.95386662 0.95498503 0.9555239 0.95548083 0.9551674 0.95442758 0.95327341 0.95194429 0.95041866 0.9484181 0.94839027 0.94838056 0.94813039 0.94753901 0.94645196 0.94504703 0.9433144 0.94197531 0.94049427 0.93875727 0.94132922 0.9436129 0.94558364 0.94711284 0.94831802 0.94945878 0.95060227 0.95182631 0.95345901 0.95510105 0.95225316 0.95033203 0.94956701 0.94895037 0.94955832 0.95152282 0.95486528 0.95963437 0.96607045 0.97375116 0.97078626 0.96900558 0.96819786 0.96750837 0.9674355 0.96792378 0.96839035 0.96883692 0.9693902 0.96985301 0.96898817 0.96814874 0.96743874 0.96638941 0.96534457 0.96425351 0.96281969 0.96155971 0.96053251 0.95926107 0.95959913 0.96025143 0.96154033 0.96262988 0.96359875 0.96467154 0.9654114 0.96625109 0.96744643 0.96878244 0.97046916 0.97234778 0.97422228 0.9759326 0.97713045 0.97892333 0.98039008 0.98147316 0.98248415 0.9832885 0.98458694 0.9866985 0.98911057 0.99119702 0.99315819 0.99524701 0.99668121 0.99826512 0.99959594 1 0.99866374 0.99576531 0.99062502 0.98155399 0.96876193 0.95174168 0.92942389 0.90240499 0.8705554 0.83207693 0.78626172 0.73649625 0.68202115 0.62329799 0.56418429 0.50680538 0.4515209 0.4006232 0.35430248 0.30995212 0.26941268 0.23418861 0.20232924 0.17373524 0.14941215 0.1290062 0.11157271 0.09693492 0.08490613 0.07409041 0.06449244 0.056343 0.0493999 0.04321207 0.03800099 0.03348242 0.02900748 0.0251013 0.02003645 0.01384563 0.00850471 0.00443484 0.00085324', 'units': 'nm', 'wavelength_max': 2303.0, 'wavelength_min': 2065.0}, 'b8a': {'bandwidth': 20.0, 'central_wavelength': 864.0, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7608204', 'physical_gain': '5.76107136', 'spectral_response_step': '1', 'spectral_response_values': '0.00167523 0.01602231 0.03225867 0.07345268 0.1689243 0.34543042 0.56923369 0.79611745 0.93749188 0.98102805 0.98742384 0.99457226 0.99912415 0.99993652 1 0.99437257 0.98756135 0.98263615 0.9790323 0.97397518 0.97130259 0.9645338 0.95610202 0.93941552 0.89155652 0.77601041 0.5951886 0.37588812 0.18394037 0.07870072 0.0332686 0.01575167 0.00159818', 'units': 'nm', 'wavelength_max': 880.0, 'wavelength_min': 848.0}}, 'declared_accuracy_of_AOT_model': 0.0, 'declared_accuracy_of_radiative_transfer_model': 0.0, 'declared_accuracy_of_water_vapour_model': 0.0, 'electronic_crosstalk_correction_flag': 'null', 'eopf_category': 'eoproduct', 'geometric_refinement': {'mean_value_of_residual_displacements_at_all_tie_points_after_refinement_m': {'x_mean': 'null', 'y_mean': 'null'}, 'spacecraft_rotation': {'X': {'coefficients': 'null', 'degree': 'null'}, 'Y': {'coefficients': 'null', 'degree': 'null'}, 'Z': {'coefficients': 'null', 'degree': 'null'}}, 'standard_deviation_of_residual_displacements_at_all_tie_points_after_refinement_m': {'x_stdv': 'null', 'y_stdv': 'null'}}, 'history': [{'output': 'Downlinked Stream', 'type': 'Raw Data'}, {'inputs': 'Downlinked Stream', 'organisation': 'ESA', 'output': 'S2MSIL0__etc', 'processor': 'L0', 'type': 'Level-0 Product'}, {'inputs': {'Level-0 Product': 'S2MSIL0__etc', 'list of used processing parameters file names': 'S2B_OPER_GIP_PROBAS_MPC__20240717T000511_V20240723T070000_21000101T000000_B00,S2B_OPER_GIP_ATMIMA_MPC__20170206T103051_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_INVLOC_MPC__20170523T080300_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_LREXTR_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_OLQCPA_MPC__20250210T000044_V20250211T000000_21000101T000000_B00,S2B_OPER_GIP_ATMSAD_MPC__20170324T155501_V20170306T000000_21000101T000000_B00,S2B_OPER_GIP_BLINDP_MPC__20170221T000000_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_CLOINV_MPC__20210609T000002_V20210823T030000_21000101T000000_B00,S2B_OPER_GIP_CLOPAR_MPC__20220120T000001_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_CONVER_MPC__20150710T131444_V20150627T000000_21000101T000000_B00,S2B_OPER_GIP_DATATI_MPC__20170428T123038_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_DECOMP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2__OPER_GIP_EARMOD_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_ECMWFP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2B_OPER_GIP_G2PARA_MPC__20250128T000031_V20250130T001500_21000101T000000_B00,S2B_OPER_GIP_G2PARE_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_GEOPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_HRTPAR_MPC__20221206T000000_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_INTDET_MPC__20220120T000010_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_JP2KPA_MPC__20220120T000006_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_MASPAR_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_PRDLOC_MPC__20180301T130000_V20180305T014000_21000101T000000_B00,S2B_OPER_GIP_R2ABCA_MPC__20250512T170000_V20250514T010000_21000101T000000_B00,S2B_OPER_GIP_R2BINN_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_R2CRCO_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEPI_MPC__20250312T000008_V20250313T000000_21000101T000000_B00,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B12,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B03,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B05,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B08,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B04,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B10,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B01,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B06,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B09,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B02,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B8A,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B07,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B06,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B02,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B01,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B8A,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B08,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B05,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B09,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B04,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B12,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B03,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B07,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2NOMO_MPC__20250117T000002_V20250121T080000_21000101T000000_B00,S2B_OPER_GIP_R2PARA_MPC__20221206T000009_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_R2SWIR_MPC__20170523T080300_V20170517T090600_21000101T000000_B00,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_RESPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_SPAMOD_MPC__20250428T000037_V20250430T000000_21000101T000000_B00,S2B_OPER_GIP_TILPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B8A,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B01,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B11,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B08,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B03,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B07,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B02,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B12,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B04,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B06,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B09,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B05,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B10,S2__OPER_GIP_L2ACSC_MPC__20220121T000003_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_L2ACAC_MPC__20220121T000004_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_PROBA2_MPC__20240809T000511_V20240813T070000_21000101T000000_B00', 'used CAMS file names': 'S2__OPER_AUX_CAMSFO_ADG__20250518T000000_V20250518T000000_20250520T010000', 'used DEM file name': 'CopernicusDEM30', 'used ECMWF file names': 'S2__OPER_AUX_ECMWFD_ADG__20250518T000000_V20250518T090000_20250520T030000', 'used GRI file name': 'S2A_OPER_GRI_MSIL1B_MPC__20160522T000734_S20160210T113515,S2A_OPER_GRI_MSIL1B_MPC__20160630T162915_S20160609T113147,S2A_OPER_GRI_MSIL1B_MPC__20160910T073127_S20160311T113330', 'used IERS file name': 'S2__OPER_AUX_UT1UTC_PDMC_20250515T000000_V20250516T000000_20260515T000000'}, 'organisation': 'ESA', 'output': 'S2MSIL1A_etc', 'processor': 'Sentinel-2 IPF', 'type': 'Level-1A Product', 'version': '???'}, {'inputs': {'Level-1A Product': 'S2MSIL1A_etc', 'list of used processing parameters file names': 'S2B_OPER_GIP_PROBAS_MPC__20240717T000511_V20240723T070000_21000101T000000_B00,S2B_OPER_GIP_ATMIMA_MPC__20170206T103051_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_INVLOC_MPC__20170523T080300_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_LREXTR_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_OLQCPA_MPC__20250210T000044_V20250211T000000_21000101T000000_B00,S2B_OPER_GIP_ATMSAD_MPC__20170324T155501_V20170306T000000_21000101T000000_B00,S2B_OPER_GIP_BLINDP_MPC__20170221T000000_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_CLOINV_MPC__20210609T000002_V20210823T030000_21000101T000000_B00,S2B_OPER_GIP_CLOPAR_MPC__20220120T000001_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_CONVER_MPC__20150710T131444_V20150627T000000_21000101T000000_B00,S2B_OPER_GIP_DATATI_MPC__20170428T123038_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_DECOMP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2__OPER_GIP_EARMOD_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_ECMWFP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2B_OPER_GIP_G2PARA_MPC__20250128T000031_V20250130T001500_21000101T000000_B00,S2B_OPER_GIP_G2PARE_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_GEOPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_HRTPAR_MPC__20221206T000000_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_INTDET_MPC__20220120T000010_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_JP2KPA_MPC__20220120T000006_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_MASPAR_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_PRDLOC_MPC__20180301T130000_V20180305T014000_21000101T000000_B00,S2B_OPER_GIP_R2ABCA_MPC__20250512T170000_V20250514T010000_21000101T000000_B00,S2B_OPER_GIP_R2BINN_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_R2CRCO_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEPI_MPC__20250312T000008_V20250313T000000_21000101T000000_B00,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B12,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B03,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B05,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B08,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B04,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B10,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B01,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B06,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B09,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B02,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B8A,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B07,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B06,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B02,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B01,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B8A,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B08,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B05,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B09,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B04,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B12,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B03,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B07,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2NOMO_MPC__20250117T000002_V20250121T080000_21000101T000000_B00,S2B_OPER_GIP_R2PARA_MPC__20221206T000009_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_R2SWIR_MPC__20170523T080300_V20170517T090600_21000101T000000_B00,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_RESPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_SPAMOD_MPC__20250428T000037_V20250430T000000_21000101T000000_B00,S2B_OPER_GIP_TILPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B8A,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B01,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B11,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B08,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B03,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B07,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B02,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B12,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B04,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B06,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B09,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B05,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B10,S2__OPER_GIP_L2ACSC_MPC__20220121T000003_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_L2ACAC_MPC__20220121T000004_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_PROBA2_MPC__20240809T000511_V20240813T070000_21000101T000000_B00', 'used CAMS file names': 'S2__OPER_AUX_CAMSFO_ADG__20250518T000000_V20250518T000000_20250520T010000', 'used DEM file name': 'CopernicusDEM30', 'used ECMWF file names': 'S2__OPER_AUX_ECMWFD_ADG__20250518T000000_V20250518T090000_20250520T030000', 'used GRI file name': 'S2A_OPER_GRI_MSIL1B_MPC__20160522T000734_S20160210T113515,S2A_OPER_GRI_MSIL1B_MPC__20160630T162915_S20160609T113147,S2A_OPER_GRI_MSIL1B_MPC__20160910T073127_S20160311T113330', 'used IERS file name': 'S2__OPER_AUX_UT1UTC_PDMC_20250515T000000_V20250516T000000_20260515T000000'}, 'organisation': 'ESA', 'output': 'S2MSIL1B_etc', 'processor': 'Sentinel-2 IPF', 'type': 'Level-1B Product', 'version': '???'}, {'inputs': {'Level-1B Product': 'S2MSIL1B_etc', 'list of used processing parameters file names': 'S2B_OPER_GIP_PROBAS_MPC__20240717T000511_V20240723T070000_21000101T000000_B00,S2B_OPER_GIP_ATMIMA_MPC__20170206T103051_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_INVLOC_MPC__20170523T080300_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_LREXTR_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_OLQCPA_MPC__20250210T000044_V20250211T000000_21000101T000000_B00,S2B_OPER_GIP_ATMSAD_MPC__20170324T155501_V20170306T000000_21000101T000000_B00,S2B_OPER_GIP_BLINDP_MPC__20170221T000000_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_CLOINV_MPC__20210609T000002_V20210823T030000_21000101T000000_B00,S2B_OPER_GIP_CLOPAR_MPC__20220120T000001_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_CONVER_MPC__20150710T131444_V20150627T000000_21000101T000000_B00,S2B_OPER_GIP_DATATI_MPC__20170428T123038_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_DECOMP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2__OPER_GIP_EARMOD_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_ECMWFP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2B_OPER_GIP_G2PARA_MPC__20250128T000031_V20250130T001500_21000101T000000_B00,S2B_OPER_GIP_G2PARE_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_GEOPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_HRTPAR_MPC__20221206T000000_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_INTDET_MPC__20220120T000010_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_JP2KPA_MPC__20220120T000006_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_MASPAR_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_PRDLOC_MPC__20180301T130000_V20180305T014000_21000101T000000_B00,S2B_OPER_GIP_R2ABCA_MPC__20250512T170000_V20250514T010000_21000101T000000_B00,S2B_OPER_GIP_R2BINN_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_R2CRCO_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEPI_MPC__20250312T000008_V20250313T000000_21000101T000000_B00,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B12,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B03,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B05,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B08,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B04,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B10,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B01,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B06,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B09,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B02,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B8A,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B07,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B06,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B02,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B01,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B8A,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B08,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B05,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B09,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B04,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B12,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B03,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B07,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2NOMO_MPC__20250117T000002_V20250121T080000_21000101T000000_B00,S2B_OPER_GIP_R2PARA_MPC__20221206T000009_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_R2SWIR_MPC__20170523T080300_V20170517T090600_21000101T000000_B00,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_RESPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_SPAMOD_MPC__20250428T000037_V20250430T000000_21000101T000000_B00,S2B_OPER_GIP_TILPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B8A,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B01,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B11,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B08,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B03,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B07,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B02,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B12,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B04,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B06,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B09,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B05,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B10,S2__OPER_GIP_L2ACSC_MPC__20220121T000003_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_L2ACAC_MPC__20220121T000004_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_PROBA2_MPC__20240809T000511_V20240813T070000_21000101T000000_B00', 'used CAMS file names': 'S2__OPER_AUX_CAMSFO_ADG__20250518T000000_V20250518T000000_20250520T010000', 'used DEM file name': 'CopernicusDEM30', 'used ECMWF file names': 'S2__OPER_AUX_ECMWFD_ADG__20250518T000000_V20250518T090000_20250520T030000', 'used GRI file name': 'S2A_OPER_GRI_MSIL1B_MPC__20160522T000734_S20160210T113515,S2A_OPER_GRI_MSIL1B_MPC__20160630T162915_S20160609T113147,S2A_OPER_GRI_MSIL1B_MPC__20160910T073127_S20160311T113330', 'used IERS file name': 'S2__OPER_AUX_UT1UTC_PDMC_20250515T000000_V20250516T000000_20260515T000000'}, 'organisation': 'ESA', 'output': 'S2MSIL1C_etc', 'processor': 'Sentinel-2 IPF', 'type': 'Level-1C Product', 'version': '???'}, {'inputs': {'Level-1C Product': 'S2MSIL1C_etc', 'list of used processing parameters file names': 'S2B_OPER_GIP_PROBAS_MPC__20240717T000511_V20240723T070000_21000101T000000_B00,S2B_OPER_GIP_ATMIMA_MPC__20170206T103051_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_INVLOC_MPC__20170523T080300_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_LREXTR_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_OLQCPA_MPC__20250210T000044_V20250211T000000_21000101T000000_B00,S2B_OPER_GIP_ATMSAD_MPC__20170324T155501_V20170306T000000_21000101T000000_B00,S2B_OPER_GIP_BLINDP_MPC__20170221T000000_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_CLOINV_MPC__20210609T000002_V20210823T030000_21000101T000000_B00,S2B_OPER_GIP_CLOPAR_MPC__20220120T000001_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_CONVER_MPC__20150710T131444_V20150627T000000_21000101T000000_B00,S2B_OPER_GIP_DATATI_MPC__20170428T123038_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_DECOMP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2__OPER_GIP_EARMOD_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_ECMWFP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2B_OPER_GIP_G2PARA_MPC__20250128T000031_V20250130T001500_21000101T000000_B00,S2B_OPER_GIP_G2PARE_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_GEOPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_HRTPAR_MPC__20221206T000000_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_INTDET_MPC__20220120T000010_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_JP2KPA_MPC__20220120T000006_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_MASPAR_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_PRDLOC_MPC__20180301T130000_V20180305T014000_21000101T000000_B00,S2B_OPER_GIP_R2ABCA_MPC__20250512T170000_V20250514T010000_21000101T000000_B00,S2B_OPER_GIP_R2BINN_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_R2CRCO_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEPI_MPC__20250312T000008_V20250313T000000_21000101T000000_B00,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B12,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B03,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B05,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B08,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B04,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B10,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B01,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B06,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B09,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B02,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B8A,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B07,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B06,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B02,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B01,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B8A,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B08,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B05,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B09,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B04,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B12,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B03,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B07,S2B_OPER_GIP_R2EQOG_MPC__20250512T170000_V20250514T010000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2NOMO_MPC__20250117T000002_V20250121T080000_21000101T000000_B00,S2B_OPER_GIP_R2PARA_MPC__20221206T000009_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_R2SWIR_MPC__20170523T080300_V20170517T090600_21000101T000000_B00,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_RESPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_SPAMOD_MPC__20250428T000037_V20250430T000000_21000101T000000_B00,S2B_OPER_GIP_TILPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B8A,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B01,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B11,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B08,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B03,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B07,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B02,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B12,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B04,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B06,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B09,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B05,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B10,S2__OPER_GIP_L2ACSC_MPC__20220121T000003_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_L2ACAC_MPC__20220121T000004_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_PROBA2_MPC__20240809T000511_V20240813T070000_21000101T000000_B00', 'used CAMS file names': 'S2__OPER_AUX_CAMSFO_ADG__20250518T000000_V20250518T000000_20250520T010000', 'used DEM file name': 'CopernicusDEM30', 'used ECMWF file names': 'S2__OPER_AUX_ECMWFD_ADG__20250518T000000_V20250518T090000_20250520T030000', 'used ESA CCI land cover map file name': 'ESACCI-LC-L4-LCCS-Map-300m-P1Y-2015-v2.0.7.tif', 'used ESA CCI snow condition map folder name': 'ESACCI-LC-L4-Snow-Cond-500m-MONTHLY-2000-2012-v2.4', 'used ESA CCI water bodies map file name': 'ESACCI-LC-L4-WB-Map-150m-P13Y-2000-v4.0.tif', 'used GRI file name': 'S2A_OPER_GRI_MSIL1B_MPC__20160522T000734_S20160210T113515,S2A_OPER_GRI_MSIL1B_MPC__20160630T162915_S20160609T113147,S2A_OPER_GRI_MSIL1B_MPC__20160910T073127_S20160311T113330', 'used IERS file name': 'S2__OPER_AUX_UT1UTC_PDMC_20250515T000000_V20250516T000000_20260515T000000', 'used LibRadTran Look-Up Tables list': 'None', 'used snow climatology map file name': 'GlobalSnowMap.tiff'}, 'organisation': 'ESA', 'output': 'S2MSIL2A_etc', 'processor': 'Sentinel-2 IPF', 'type': 'Level-2A Product', 'version': '???'}], 'horizontal_CRS_code': 'EPSG:32629', 'horizontal_CRS_name': 'WGS84 / UTM zone 29N', 'mean_sensing_time': '2025-05-18T11:33:44.731983Z', 'mean_sun_azimuth_angle_in_deg_for_all_bands_all_detectors': 114.555334769026, 'mean_sun_zenith_angle_in_deg_for_all_bands_all_detectors': 16.7208464648829, 'mean_value_of_aerosol_optical_thickness': 0.308178, 'mean_value_of_total_water_vapour_content': 1.11232, 'meteo': {'source': 'ECMWF', 'type': 'FORECAST'}, 'multispectral_registration_assessment': 'N/A', 'onboard_compression_flag': 'true', 'onboard_equalization_flag': 'null', 'optical_crosstalk_correction_flag': 'null', 'ozone_source': 'AUX_ECMWFT', 'ozone_value': 311.909424, 'percentage_of_degraded_MSI_data': 0.0, 'planimetric_stability_assessment_from_AOCS': '\n ', 'product_quality_status': 'PASSED,PASSED,PASSED,PASSED,PASSED,PASSED', 'reflectance_correction_factor_from_the_Sun-Earth_distance_variation_computed_using_the_acquisition_date': 0.979070847619781, 'spectral_band_of_reference': 'N/A'}
- stac_discovery :
- {'assets': {'analytic': {'eo:bands': [{'center_wavelength': 0.4423, 'common_name': 'coastal', 'full_width_half_max': 0.02, 'name': '01', 'solar_illumination': '1874.3'}, {'center_wavelength': 0.4923, 'common_name': 'blue', 'full_width_half_max': 0.065, 'name': '02', 'solar_illumination': '1959.75'}, {'center_wavelength': 0.559, 'common_name': 'green', 'full_width_half_max': 0.035, 'name': '03', 'solar_illumination': '1824.93'}, {'center_wavelength': 0.665, 'common_name': 'red', 'full_width_half_max': 0.03, 'name': '04', 'solar_illumination': '1512.79'}, {'center_wavelength': 0.7038, 'full_width_half_max': 0.015, 'name': '05', 'solar_illumination': '1425.78'}, {'center_wavelength': 0.7391000000000001, 'full_width_half_max': 0.015, 'name': '06', 'solar_illumination': '1291.13'}, {'center_wavelength': 0.7797000000000001, 'full_width_half_max': 0.02, 'name': '07', 'solar_illumination': '1175.57'}, {'center_wavelength': 0.833, 'common_name': 'nir', 'full_width_half_max': 0.105, 'name': '08', 'solar_illumination': '1041.28'}, {'center_wavelength': 0.864, 'full_width_half_max': 0.02, 'name': '8A', 'solar_illumination': '953.93'}, {'center_wavelength': 0.9432, 'full_width_half_max': 0.02, 'name': '09', 'solar_illumination': '817.58'}, {'center_wavelength': 1.3769, 'common_name': 'cirrus', 'full_width_half_max': 0.03, 'name': '10', 'solar_illumination': '365.41'}, {'center_wavelength': 1.6104, 'common_name': 'swir16', 'full_width_half_max': 0.09, 'name': '11', 'solar_illumination': '247.08'}, {'center_wavelength': 2.1856999999999998, 'common_name': 'swir22', 'full_width_half_max': 0.18, 'name': '12', 'solar_illumination': '87.75'}], 'eo:cloud_cover': 0.009198, 'eo:snow_cover': 0.0, 'href': 'null'}}, 'bbox': [-9.913869037537504, 27.019735585570864, -11.034076356260641, 28.022372192571748], 'geometry': {'coordinates': [[[-9.913869037537504, 27.576269024486606], [-9.928780222747973, 27.52232000320648], [-9.969472881615348, 27.3747446613974], [-10.010091332532475, 27.22717356028996], [-10.050337008816811, 27.079470773025964], [-10.063951518262359, 27.029627391642432], [-11.015926461321431, 27.019735585570864], [-11.034076356260641, 28.01038067774639], [-9.917556498778872, 28.022372192571748], [-9.913869037537504, 27.576269024486606]]], 'type': 'Polygon'}, 'id': 'S2B_MSIL2A_20250518T112119_N0511_R037_T29RLL_20250518T140519.SAFE', 'links': [{'href': './.zattrs.json', 'rel': 'self', 'type': 'application/json'}], 'properties': {'bands': [{'center_wavelength': 442.3, 'common_name': 'coastal', 'full_width_half_max': 0.02, 'name': 'b01', 'solar_illumination': 1874.3}, {'center_wavelength': 492.3, 'common_name': 'blue', 'full_width_half_max': 0.065, 'name': 'b02', 'solar_illumination': 1959.75}, {'center_wavelength': 559.0, 'common_name': 'green', 'full_width_half_max': 0.035, 'name': 'b03', 'solar_illumination': 1824.93}, {'center_wavelength': 665.0, 'common_name': 'red', 'full_width_half_max': 0.03, 'name': 'b04', 'solar_illumination': 1512.79}, {'center_wavelength': 703.8, 'common_name': 'rededge', 'full_width_half_max': 0.015, 'name': 'b05', 'solar_illumination': 1425.78}, {'center_wavelength': 739.1, 'common_name': 'rededge', 'full_width_half_max': 0.015, 'name': 'b06', 'solar_illumination': 1291.13}, {'center_wavelength': 779.7, 'common_name': 'rededge', 'full_width_half_max': 0.02, 'name': 'b07', 'solar_illumination': 1175.57}, {'center_wavelength': 833.0, 'common_name': 'nir', 'full_width_half_max': 0.105, 'name': 'b08', 'solar_illumination': 1041.28}, {'center_wavelength': 864.0, 'common_name': 'nir08', 'full_width_half_max': 0.02, 'name': 'b8a', 'solar_illumination': 953.93}, {'center_wavelength': 943.2, 'common_name': 'nir09', 'full_width_half_max': 0.02, 'name': 'b09', 'solar_illumination': 817.58}, {'center_wavelength': 1376.9, 'common_name': 'cirrus', 'full_width_half_max': 0.03, 'name': 'b10', 'solar_illumination': 365.41}, {'center_wavelength': 1610.4, 'common_name': 'swir16', 'full_width_half_max': 0.09, 'name': 'b11', 'solar_illumination': 247.08}, {'center_wavelength': 2185.7, 'common_name': 'swir22', 'full_width_half_max': 0.18, 'name': 'b12', 'solar_illumination': 87.75}], 'constellation': 'sentinel-2', 'created': '2025-05-18T14:05:19+00:00', 'datetime': None, 'end_datetime': '2025-05-18T11:21:19.024000+00:00', 'eo:cloud_cover': 0.009198, 'eo:snow_cover': 0.0, 'eopf:baseline': '05.11', 'eopf:data_take_id': 'GS2B_20250518T112119_042820_N05.11', 'eopf:instrument_mode': 'INS-NOBS', 'eopf:resolutions': {'bands 01, 09, 10': '60', 'bands 02, 03, 04, 08': '10', 'bands 05, 06, 07, 8A, 11, 12': '20'}, 'instrument': 'msi', 'mission': 'copernicus', 'platform': 'sentinel-2b', 'processing:expression': 'systematic', 'processing:facility': 'ESA', 'processing:level': 'L2A', 'processing:lineage': 'IPF L2A processor', 'processing:software': {'Sentinel-2 IPF': ' '}, 'processing:version': '', 'product:timeline': 'NRT', 'product:timeliness': 'PT3H', 'product:timeliness_category': 'NRT', 'product:type': 'S02MSIL2A', 'proj:bbox': [300000.0, 2990220.0, 409800.0, 3100020.0], 'proj:epsg': 32629, 'providers': [{'name': 'L2A Processor', 'roles': ['processor']}, {'name': 'ESA', 'roles': ['producer']}], 'sat:absolute_orbit': 42820, 'sat:orbit_state': 'descending', 'sat:platform_international_designator': '2015-028A', 'sat:relative_orbit': 37, 'sci:doi': '10.5270/S2_-znk9xsj', 'start_datetime': '2025-05-18T11:21:19.024000+00:00'}, 'stac_extensions': ['https://stac-extensions.github.io/eopf/v1.0.0/schema.json', 'https://stac-extensions.github.io/eo/v1.1.0/schema.json', 'https://stac-extensions.github.io/sat/v1.0.0/schema.json', 'https://stac-extensions.github.io/view/v1.0.0/schema.json', 'https://stac-extensions.github.io/scientific/v1.0.0/schema.json', 'https://stac-extensions.github.io/processing/v1.2.0/schema.json', 'https://stac-extensions.github.io/product/v0.1.0/schema.json'], 'stac_version': '1.0.0', 'type': 'Feature'}
What is xarray?#
Xarray introduces labels in the form of dimensions, coordinates and attributes on top of raw NumPy-like multi-dimensional arrays, which allows for a more intuitive, more concise, and less error-prone developer experience.
How is xarray structured?#
Xarray has two core data structures, which build upon and extend the core strengths of NumPy and Pandas libraries. Both data structures are fundamentally N-dimensional:
DataTree is a tree-like hierarchical collection of xarray objects;
DataArray is the implementation of a labeled, N-dimensional array. It is an N-D generalization of a Pandas.Series. The name DataArray itself is borrowed from Fernando Perez’s datarray project, which prototyped a similar data structure.
Dataset is a multi-dimensional, in-memory array database. It is a dict-like container of DataArray objects aligned along any number of shared dimensions, and serves a similar purpose in xarray as the pandas.DataFrame.
Accessing Coordinates and Data Variables#
DataArray, within Datasets, can be accessed through:
the dot notation like
Dataset.NameofVariableor using square brackets, like Dataset[‘NameofVariable’] (NameofVariable needs to be a string so use quotes or double quotes)
ds["quality/l2a_quicklook/r60m/tci"]
<xarray.DataArray 'tci' (band: 3, y: 1830, x: 1830)> Size: 10MB dask.array<open_dataset-tci, shape=(3, 1830, 1830), dtype=uint8, chunksize=(1, 305, 305), chunktype=numpy.ndarray> Coordinates: * band (band) int64 24B 1 2 3 * x (x) int64 15kB 300030 300090 300150 300210 ... 409650 409710 409770 * y (y) int64 15kB 3099990 3099930 3099870 ... 2990370 2990310 2990250 Attributes: (6)
- band: 3
- y: 1830
- x: 1830
- dask.array<chunksize=(1, 305, 305), meta=np.ndarray>
Array Chunk Bytes 9.58 MiB 90.84 kiB Shape (3, 1830, 1830) (1, 305, 305) Dask graph 108 chunks in 2 graph layers Data type uint8 numpy.ndarray - band(band)int641 2 3
array([1, 2, 3])
- x(x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- y(y)int643099990 3099930 ... 2990310 2990250
array([3099990, 3099930, 3099870, ..., 2990370, 2990310, 2990250])
- bandPandasIndex
PandasIndex(Index([1, 2, 3], dtype='int64', name='band'))
- xPandasIndex
PandasIndex(Index([300030, 300090, 300150, 300210, 300270, 300330, 300390, 300450, 300510, 300570, ... 409230, 409290, 409350, 409410, 409470, 409530, 409590, 409650, 409710, 409770], dtype='int64', name='x', length=1830)) - yPandasIndex
PandasIndex(Index([3099990, 3099930, 3099870, 3099810, 3099750, 3099690, 3099630, 3099570, 3099510, 3099450, ... 2990790, 2990730, 2990670, 2990610, 2990550, 2990490, 2990430, 2990370, 2990310, 2990250], dtype='int64', name='y', length=1830))
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
ds.quality.l2a_quicklook.r60m.tci
<xarray.DataArray 'tci' (band: 3, y: 1830, x: 1830)> Size: 10MB dask.array<open_dataset-tci, shape=(3, 1830, 1830), dtype=uint8, chunksize=(1, 305, 305), chunktype=numpy.ndarray> Coordinates: * band (band) int64 24B 1 2 3 * x (x) int64 15kB 300030 300090 300150 300210 ... 409650 409710 409770 * y (y) int64 15kB 3099990 3099930 3099870 ... 2990370 2990310 2990250 Attributes: (6)
- band: 3
- y: 1830
- x: 1830
- dask.array<chunksize=(1, 305, 305), meta=np.ndarray>
Array Chunk Bytes 9.58 MiB 90.84 kiB Shape (3, 1830, 1830) (1, 305, 305) Dask graph 108 chunks in 2 graph layers Data type uint8 numpy.ndarray - band(band)int641 2 3
array([1, 2, 3])
- x(x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- y(y)int643099990 3099930 ... 2990310 2990250
array([3099990, 3099930, 3099870, ..., 2990370, 2990310, 2990250])
- bandPandasIndex
PandasIndex(Index([1, 2, 3], dtype='int64', name='band'))
- xPandasIndex
PandasIndex(Index([300030, 300090, 300150, 300210, 300270, 300330, 300390, 300450, 300510, 300570, ... 409230, 409290, 409350, 409410, 409470, 409530, 409590, 409650, 409710, 409770], dtype='int64', name='x', length=1830)) - yPandasIndex
PandasIndex(Index([3099990, 3099930, 3099870, 3099810, 3099750, 3099690, 3099630, 3099570, 3099510, 3099450, ... 2990790, 2990730, 2990670, 2990610, 2990550, 2990490, 2990430, 2990370, 2990310, 2990250], dtype='int64', name='y', length=1830))
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Accessing metadata#
Metadata describes the data itself — offering context, provenance, and characteristics — in a way that can be read by both humans and machines.
ds["quality/l2a_quicklook/r60m/tci"].attrs
{'_eopf_attrs': {'coordinates': ['band', 'x', 'y'],
'dimensions': ['band', 'y', 'x']},
'proj:bbox': [300000.0, 2990220.0, 409800.0, 3100020.0],
'proj:epsg': 32629,
'proj:shape': [1830, 1830],
'proj:transform': [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0],
'proj:wkt2': 'PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]'}
Tip
Metadata everywhere! In the command above, we are showing the metadata (attributes) of one variable called “quality/l2a_quicklook/r60m/tci”. As an exercise, try the command:
ds.attrs
What do you observe?
Quick Visualisation#
As an example, we plot the quicklook image to view the RGB image.
import matplotlib.colors as mcolors
import matplotlib.pyplot as plt
ds.quality.l2a_quicklook.r60m.tci.plot.imshow(rgb="band")
<matplotlib.image.AxesImage at 0x167d99cd0>
Selection methods#
As underneath DataArrays are Numpy Array objects (that implement the standard Python x[obj] (x: array, obj: int,slice) syntax). Their data can be accessed through the same approach of numpy indexing.
ds["quality/l2a_quicklook/r60m/tci"][0,0,0]
<xarray.DataArray 'tci' ()> Size: 1B
dask.array<getitem, shape=(), dtype=uint8, chunksize=(), chunktype=numpy.ndarray>
Coordinates:
band int64 8B 1
x int64 8B 300030
y int64 8B 3099990
Attributes: (6)- dask.array<chunksize=(), meta=np.ndarray>
Array Chunk Bytes 1 B 1 B Shape () () Dask graph 1 chunks in 3 graph layers Data type uint8 numpy.ndarray - band()int641
array(1)
- x()int64300030
array(300030)
- y()int643099990
array(3099990)
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
or slicing
ds["quality/l2a_quicklook/r60m/tci"][0,10:110,35:150]
<xarray.DataArray 'tci' (y: 100, x: 115)> Size: 12kB
dask.array<getitem, shape=(100, 115), dtype=uint8, chunksize=(100, 115), chunktype=numpy.ndarray>
Coordinates:
band int64 8B 1
* x (x) int64 920B 302130 302190 302250 302310 ... 308850 308910 308970
* y (y) int64 800B 3099390 3099330 3099270 ... 3093570 3093510 3093450
Attributes: (6)- y: 100
- x: 115
- dask.array<chunksize=(100, 115), meta=np.ndarray>
Array Chunk Bytes 11.23 kiB 11.23 kiB Shape (100, 115) (100, 115) Dask graph 1 chunks in 3 graph layers Data type uint8 numpy.ndarray - band()int641
array(1)
- x(x)int64302130 302190 ... 308910 308970
array([302130, 302190, 302250, 302310, 302370, 302430, 302490, 302550, 302610, 302670, 302730, 302790, 302850, 302910, 302970, 303030, 303090, 303150, 303210, 303270, 303330, 303390, 303450, 303510, 303570, 303630, 303690, 303750, 303810, 303870, 303930, 303990, 304050, 304110, 304170, 304230, 304290, 304350, 304410, 304470, 304530, 304590, 304650, 304710, 304770, 304830, 304890, 304950, 305010, 305070, 305130, 305190, 305250, 305310, 305370, 305430, 305490, 305550, 305610, 305670, 305730, 305790, 305850, 305910, 305970, 306030, 306090, 306150, 306210, 306270, 306330, 306390, 306450, 306510, 306570, 306630, 306690, 306750, 306810, 306870, 306930, 306990, 307050, 307110, 307170, 307230, 307290, 307350, 307410, 307470, 307530, 307590, 307650, 307710, 307770, 307830, 307890, 307950, 308010, 308070, 308130, 308190, 308250, 308310, 308370, 308430, 308490, 308550, 308610, 308670, 308730, 308790, 308850, 308910, 308970]) - y(y)int643099390 3099330 ... 3093510 3093450
array([3099390, 3099330, 3099270, 3099210, 3099150, 3099090, 3099030, 3098970, 3098910, 3098850, 3098790, 3098730, 3098670, 3098610, 3098550, 3098490, 3098430, 3098370, 3098310, 3098250, 3098190, 3098130, 3098070, 3098010, 3097950, 3097890, 3097830, 3097770, 3097710, 3097650, 3097590, 3097530, 3097470, 3097410, 3097350, 3097290, 3097230, 3097170, 3097110, 3097050, 3096990, 3096930, 3096870, 3096810, 3096750, 3096690, 3096630, 3096570, 3096510, 3096450, 3096390, 3096330, 3096270, 3096210, 3096150, 3096090, 3096030, 3095970, 3095910, 3095850, 3095790, 3095730, 3095670, 3095610, 3095550, 3095490, 3095430, 3095370, 3095310, 3095250, 3095190, 3095130, 3095070, 3095010, 3094950, 3094890, 3094830, 3094770, 3094710, 3094650, 3094590, 3094530, 3094470, 3094410, 3094350, 3094290, 3094230, 3094170, 3094110, 3094050, 3093990, 3093930, 3093870, 3093810, 3093750, 3093690, 3093630, 3093570, 3093510, 3093450])
- xPandasIndex
PandasIndex(Index([302130, 302190, 302250, 302310, 302370, 302430, 302490, 302550, 302610, 302670, ... 308430, 308490, 308550, 308610, 308670, 308730, 308790, 308850, 308910, 308970], dtype='int64', name='x', length=115)) - yPandasIndex
PandasIndex(Index([3099390, 3099330, 3099270, 3099210, 3099150, 3099090, 3099030, 3098970, 3098910, 3098850, 3098790, 3098730, 3098670, 3098610, 3098550, 3098490, 3098430, 3098370, 3098310, 3098250, 3098190, 3098130, 3098070, 3098010, 3097950, 3097890, 3097830, 3097770, 3097710, 3097650, 3097590, 3097530, 3097470, 3097410, 3097350, 3097290, 3097230, 3097170, 3097110, 3097050, 3096990, 3096930, 3096870, 3096810, 3096750, 3096690, 3096630, 3096570, 3096510, 3096450, 3096390, 3096330, 3096270, 3096210, 3096150, 3096090, 3096030, 3095970, 3095910, 3095850, 3095790, 3095730, 3095670, 3095610, 3095550, 3095490, 3095430, 3095370, 3095310, 3095250, 3095190, 3095130, 3095070, 3095010, 3094950, 3094890, 3094830, 3094770, 3094710, 3094650, 3094590, 3094530, 3094470, 3094410, 3094350, 3094290, 3094230, 3094170, 3094110, 3094050, 3093990, 3093930, 3093870, 3093810, 3093750, 3093690, 3093630, 3093570, 3093510, 3093450], dtype='int64', name='y'))
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
As it is not easy to remember the order of dimensions, Xarray really helps by making it possible to select the position using names:
.isel -> selection based on positional index
.sel -> selection based on coordinate values
We first check the number of elements in each coordinate of the Data Variable using the built-in method sizes. Same result can be achieved querying each coordinate using the Python built-in function len.
print(ds["quality/l2a_quicklook/r60m/tci"].sizes)
Frozen({'band': 3, 'y': 1830, 'x': 1830})
ds["quality/l2a_quicklook/r60m/tci"].isel(band=0, y=100, x=10)
<xarray.DataArray 'tci' ()> Size: 1B
dask.array<getitem, shape=(), dtype=uint8, chunksize=(), chunktype=numpy.ndarray>
Coordinates:
band int64 8B 1
x int64 8B 300630
y int64 8B 3093990
Attributes: (6)- dask.array<chunksize=(), meta=np.ndarray>
Array Chunk Bytes 1 B 1 B Shape () () Dask graph 1 chunks in 3 graph layers Data type uint8 numpy.ndarray - band()int641
array(1)
- x()int64300630
array(300630)
- y()int643093990
array(3093990)
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Or a slice
ds["quality/l2a_quicklook/r60m/tci"].isel(band=0, y=slice(10,100), x=slice(10,100))
<xarray.DataArray 'tci' (y: 90, x: 90)> Size: 8kB
dask.array<getitem, shape=(90, 90), dtype=uint8, chunksize=(90, 90), chunktype=numpy.ndarray>
Coordinates:
band int64 8B 1
* x (x) int64 720B 300630 300690 300750 300810 ... 305850 305910 305970
* y (y) int64 720B 3099390 3099330 3099270 ... 3094170 3094110 3094050
Attributes: (6)- y: 90
- x: 90
- dask.array<chunksize=(90, 90), meta=np.ndarray>
Array Chunk Bytes 7.91 kiB 7.91 kiB Shape (90, 90) (90, 90) Dask graph 1 chunks in 3 graph layers Data type uint8 numpy.ndarray - band()int641
array(1)
- x(x)int64300630 300690 ... 305910 305970
array([300630, 300690, 300750, 300810, 300870, 300930, 300990, 301050, 301110, 301170, 301230, 301290, 301350, 301410, 301470, 301530, 301590, 301650, 301710, 301770, 301830, 301890, 301950, 302010, 302070, 302130, 302190, 302250, 302310, 302370, 302430, 302490, 302550, 302610, 302670, 302730, 302790, 302850, 302910, 302970, 303030, 303090, 303150, 303210, 303270, 303330, 303390, 303450, 303510, 303570, 303630, 303690, 303750, 303810, 303870, 303930, 303990, 304050, 304110, 304170, 304230, 304290, 304350, 304410, 304470, 304530, 304590, 304650, 304710, 304770, 304830, 304890, 304950, 305010, 305070, 305130, 305190, 305250, 305310, 305370, 305430, 305490, 305550, 305610, 305670, 305730, 305790, 305850, 305910, 305970]) - y(y)int643099390 3099330 ... 3094110 3094050
array([3099390, 3099330, 3099270, 3099210, 3099150, 3099090, 3099030, 3098970, 3098910, 3098850, 3098790, 3098730, 3098670, 3098610, 3098550, 3098490, 3098430, 3098370, 3098310, 3098250, 3098190, 3098130, 3098070, 3098010, 3097950, 3097890, 3097830, 3097770, 3097710, 3097650, 3097590, 3097530, 3097470, 3097410, 3097350, 3097290, 3097230, 3097170, 3097110, 3097050, 3096990, 3096930, 3096870, 3096810, 3096750, 3096690, 3096630, 3096570, 3096510, 3096450, 3096390, 3096330, 3096270, 3096210, 3096150, 3096090, 3096030, 3095970, 3095910, 3095850, 3095790, 3095730, 3095670, 3095610, 3095550, 3095490, 3095430, 3095370, 3095310, 3095250, 3095190, 3095130, 3095070, 3095010, 3094950, 3094890, 3094830, 3094770, 3094710, 3094650, 3094590, 3094530, 3094470, 3094410, 3094350, 3094290, 3094230, 3094170, 3094110, 3094050])
- xPandasIndex
PandasIndex(Index([300630, 300690, 300750, 300810, 300870, 300930, 300990, 301050, 301110, 301170, 301230, 301290, 301350, 301410, 301470, 301530, 301590, 301650, 301710, 301770, 301830, 301890, 301950, 302010, 302070, 302130, 302190, 302250, 302310, 302370, 302430, 302490, 302550, 302610, 302670, 302730, 302790, 302850, 302910, 302970, 303030, 303090, 303150, 303210, 303270, 303330, 303390, 303450, 303510, 303570, 303630, 303690, 303750, 303810, 303870, 303930, 303990, 304050, 304110, 304170, 304230, 304290, 304350, 304410, 304470, 304530, 304590, 304650, 304710, 304770, 304830, 304890, 304950, 305010, 305070, 305130, 305190, 305250, 305310, 305370, 305430, 305490, 305550, 305610, 305670, 305730, 305790, 305850, 305910, 305970], dtype='int64', name='x')) - yPandasIndex
PandasIndex(Index([3099390, 3099330, 3099270, 3099210, 3099150, 3099090, 3099030, 3098970, 3098910, 3098850, 3098790, 3098730, 3098670, 3098610, 3098550, 3098490, 3098430, 3098370, 3098310, 3098250, 3098190, 3098130, 3098070, 3098010, 3097950, 3097890, 3097830, 3097770, 3097710, 3097650, 3097590, 3097530, 3097470, 3097410, 3097350, 3097290, 3097230, 3097170, 3097110, 3097050, 3096990, 3096930, 3096870, 3096810, 3096750, 3096690, 3096630, 3096570, 3096510, 3096450, 3096390, 3096330, 3096270, 3096210, 3096150, 3096090, 3096030, 3095970, 3095910, 3095850, 3095790, 3095730, 3095670, 3095610, 3095550, 3095490, 3095430, 3095370, 3095310, 3095250, 3095190, 3095130, 3095070, 3095010, 3094950, 3094890, 3094830, 3094770, 3094710, 3094650, 3094590, 3094530, 3094470, 3094410, 3094350, 3094290, 3094230, 3094170, 3094110, 3094050], dtype='int64', name='y'))
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
The most common way to select an area (here 54x70 points) is through the labeled coordinate using the .sel method.
ds["quality/l2a_quicklook/r60m/tci"].sel(band=1, y=slice(3097890,3094710), x=slice(301830,305970))
<xarray.DataArray 'tci' (y: 54, x: 70)> Size: 4kB
dask.array<getitem, shape=(54, 70), dtype=uint8, chunksize=(54, 70), chunktype=numpy.ndarray>
Coordinates:
band int64 8B 1
* x (x) int64 560B 301830 301890 301950 302010 ... 305850 305910 305970
* y (y) int64 432B 3097890 3097830 3097770 ... 3094830 3094770 3094710
Attributes: (6)- y: 54
- x: 70
- dask.array<chunksize=(54, 70), meta=np.ndarray>
Array Chunk Bytes 3.69 kiB 3.69 kiB Shape (54, 70) (54, 70) Dask graph 1 chunks in 3 graph layers Data type uint8 numpy.ndarray - band()int641
array(1)
- x(x)int64301830 301890 ... 305910 305970
array([301830, 301890, 301950, 302010, 302070, 302130, 302190, 302250, 302310, 302370, 302430, 302490, 302550, 302610, 302670, 302730, 302790, 302850, 302910, 302970, 303030, 303090, 303150, 303210, 303270, 303330, 303390, 303450, 303510, 303570, 303630, 303690, 303750, 303810, 303870, 303930, 303990, 304050, 304110, 304170, 304230, 304290, 304350, 304410, 304470, 304530, 304590, 304650, 304710, 304770, 304830, 304890, 304950, 305010, 305070, 305130, 305190, 305250, 305310, 305370, 305430, 305490, 305550, 305610, 305670, 305730, 305790, 305850, 305910, 305970]) - y(y)int643097890 3097830 ... 3094770 3094710
array([3097890, 3097830, 3097770, 3097710, 3097650, 3097590, 3097530, 3097470, 3097410, 3097350, 3097290, 3097230, 3097170, 3097110, 3097050, 3096990, 3096930, 3096870, 3096810, 3096750, 3096690, 3096630, 3096570, 3096510, 3096450, 3096390, 3096330, 3096270, 3096210, 3096150, 3096090, 3096030, 3095970, 3095910, 3095850, 3095790, 3095730, 3095670, 3095610, 3095550, 3095490, 3095430, 3095370, 3095310, 3095250, 3095190, 3095130, 3095070, 3095010, 3094950, 3094890, 3094830, 3094770, 3094710])
- xPandasIndex
PandasIndex(Index([301830, 301890, 301950, 302010, 302070, 302130, 302190, 302250, 302310, 302370, 302430, 302490, 302550, 302610, 302670, 302730, 302790, 302850, 302910, 302970, 303030, 303090, 303150, 303210, 303270, 303330, 303390, 303450, 303510, 303570, 303630, 303690, 303750, 303810, 303870, 303930, 303990, 304050, 304110, 304170, 304230, 304290, 304350, 304410, 304470, 304530, 304590, 304650, 304710, 304770, 304830, 304890, 304950, 305010, 305070, 305130, 305190, 305250, 305310, 305370, 305430, 305490, 305550, 305610, 305670, 305730, 305790, 305850, 305910, 305970], dtype='int64', name='x')) - yPandasIndex
PandasIndex(Index([3097890, 3097830, 3097770, 3097710, 3097650, 3097590, 3097530, 3097470, 3097410, 3097350, 3097290, 3097230, 3097170, 3097110, 3097050, 3096990, 3096930, 3096870, 3096810, 3096750, 3096690, 3096630, 3096570, 3096510, 3096450, 3096390, 3096330, 3096270, 3096210, 3096150, 3096090, 3096030, 3095970, 3095910, 3095850, 3095790, 3095730, 3095670, 3095610, 3095550, 3095490, 3095430, 3095370, 3095310, 3095250, 3095190, 3095130, 3095070, 3095010, 3094950, 3094890, 3094830, 3094770, 3094710], dtype='int64', name='y'))
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 2990220.0, 409800.0, 3100020.0]
- proj:epsg :
- 32629
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 3100020.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 29N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32629"]]
Visualize on a map with a projection#
# Assign the CRS (UTM zone 32N)
# Select your DataArray slice
da = ds["quality/l2a_quicklook/r60m/tci"].sel(
band=1,
# y=slice(3097890, 3094710),
# x=slice(301830, 305970)
)
# Assign the CRS (UTM zone 32N)
da = da.rio.write_crs("EPSG:32632")
# Reproject to EPSG:4326 (PlateCarree)
da_ll = da.rio.reproject("EPSG:4326")
Plot using matplotlib and cartopy#
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
fig = plt.figure(figsize=[12, 10])
ax = plt.subplot(1, 1, 1, projection=ccrs.Mercator())
ax.coastlines(resolution='10m')
da_ll.plot(
ax=ax,
transform=ccrs.PlateCarree(),
)
plt.title("Sentinel-2 TCI (UTM 32N → Lat/Lon)", fontsize=16)
plt.show()
Warning
Limitation of Xarray 👉 You need to know where the file is!
But what if I don’t know the file?#
Earth Observation (EO) data is massive — petabytes across satellites, sensors, and time ranges.
Manually finding files? Difficult. Painful.
We need a way to search and organize EO datasets by space, time, and metadata. In short, we need a catalog.
In EO, the de-facto standard for dataset catalogs is STAC.
STAC stands for SpatioTemporal Asset Catalog. It’s an open standard, developed by the community, for describing geospatial data — especially large EO datasets like Sentinel, Landsat, or commercial imagery.
Key points:
Catalog of assets: A STAC is basically a JSON-based catalog that describes geospatial data (satellite images, derived products, etc.).
Spatiotemporal: Each dataset (called an Item) has metadata about where (bounding box, geometry) and when (timestamp) it was acquired.
Assets: The actual data files (e.g., GeoTIFFs, Cloud-optimized GeoTIFFs, NetCDF, Zarr) are linked as “assets” in the STAC Item.
Collections: Items are grouped into collections (e.g., Sentinel-2 L2A).
Catalogs: Collections can be organized hierarchically, enabling scalable discovery across petabytes of EO data.
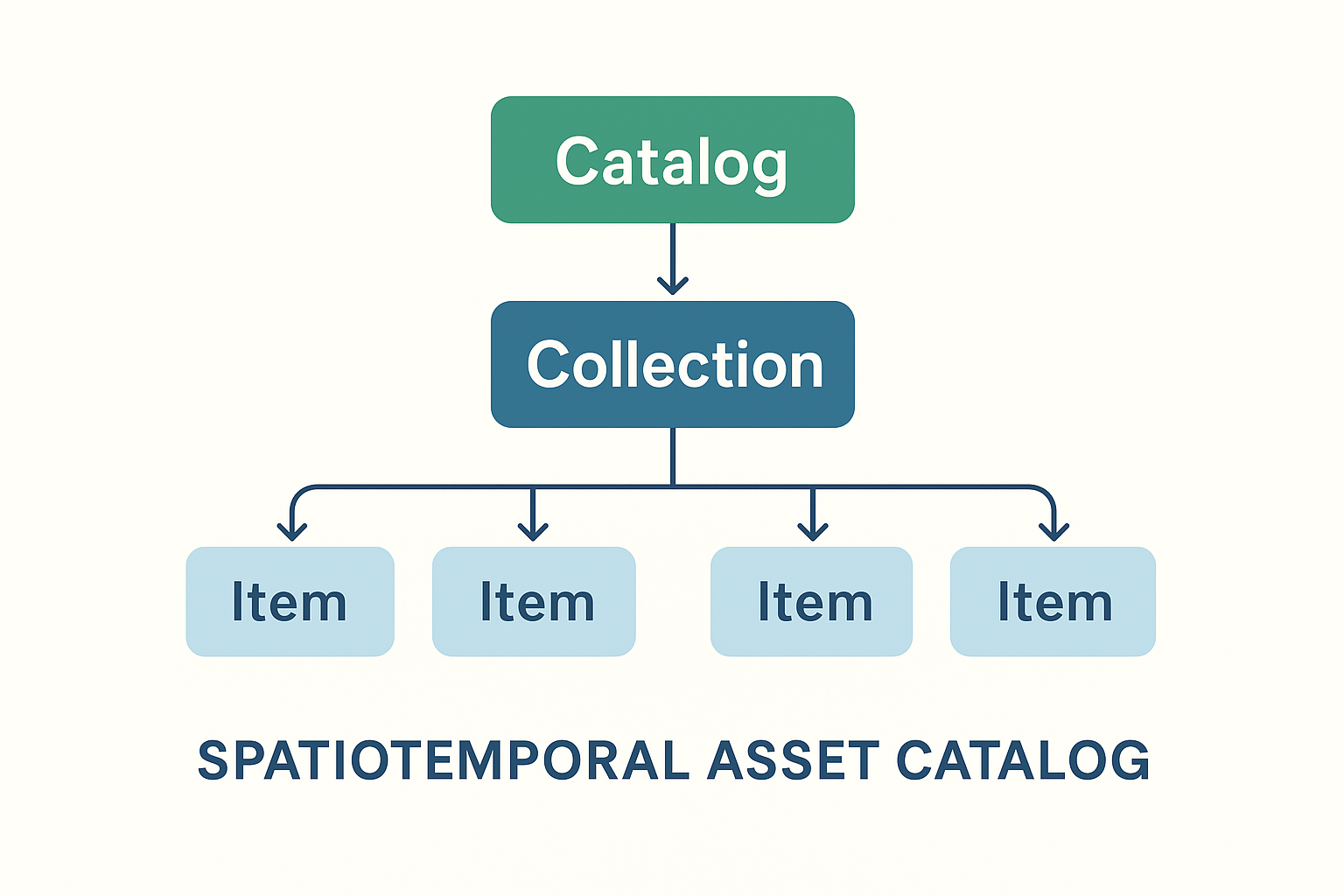
Let’s access Sentinel-2 satellite imagery using a STAC (SpatioTemporal Asset Catalog) interface. Leveraging the pystac-client, xarray, and matplotlib libraries, the notebook demonstrates how to:
Connect to a public EOPF STAC catalog hosted at https://stac.core.eopf.eodc.eu.
Perform structured searches across Sentinel-2 L2A collections, filtering by spatial extent, date range, cloud cover, and orbit characteristics.
Retrieve and load data assets directly into xarray for interactive analysis.
Visualize Sentinel-2 RGB composites along with pixel-level cloud coverage masks.
import matplotlib.colors as mcolors
import matplotlib.pyplot as plt
import numpy as np
import pystac_client
import xarray as xr
from pystac_client import CollectionSearch
from matplotlib.gridspec import GridSpec
# Initialize the collection search
search = CollectionSearch(
url="https://stac.core.eopf.eodc.eu/collections", # STAC /collections endpoint
)
# Retrieve all matching collections (as dictionaries)
for collection_dict in search.collections_as_dicts():
print(collection_dict["id"])
sentinel-2-l2a
sentinel-3-olci-l2-lfr
sentinel-1-l2-ocn
sentinel-3-slstr-l2-lst
sentinel-1-l1-grd
sentinel-2-l1c
sentinel-1-l1-slc
sentinel-3-slstr-l1-rbt
sentinel-3-olci-l1-efr
sentinel-3-olci-l1-err
sentinel-3-olci-l2-lrr
Sentinel-2 Item Search#
Querying the Sentinel-2 L2A collection by bounding box, date range, and cloud cover
catalog = pystac_client.Client.open("https://stac.core.eopf.eodc.eu")
# Search with cloud cover filter
items = list(
catalog.search(
collections=["sentinel-2-l2a"],
bbox=[7.2, 44.5, 7.4, 44.7],
datetime=["2025-01-30", "2025-05-01"],
query={"eo:cloud_cover": {"lt": 20}}, # Cloud cover less than 20%
).items()
)
print(items)
[<Item id=S2B_MSIL2A_20250430T101559_N0511_R065_T32TLQ_20250430T131328>, <Item id=S2C_MSIL2A_20250425T102041_N0511_R065_T32TLQ_20250425T155812>, <Item id=S2C_MSIL2A_20250418T103041_N0511_R108_T32TLQ_20250418T160655>, <Item id=S2C_MSIL2A_20250405T102041_N0511_R065_T32TLQ_20250405T175414>]
Quicklook Visualization for Sentinel-2#
We can use the RGB quicklook layer contained in the Sentinel-2 EOPF Zarr product for a visualization of the content:
item = items[0] # extracting the first item
ds = xr.open_dataset(
item.assets["product"].href,
**item.assets["product"].extra_fields["xarray:open_datatree_kwargs"],
) # The engine="eopf-zarr" is already embedded in the STAC metadata
ds
<xarray.Dataset> Size: 9GB
Dimensions: (
conditions_geometry_angle: 2,
conditions_geometry_band: 13,
conditions_geometry_y: 23,
conditions_geometry_x: 23,
conditions_geometry_detector: 4,
...
quality_mask_r20m_y: 5490,
quality_mask_r20m_x: 5490,
quality_mask_r60m_y: 1830,
quality_mask_r60m_x: 1830,
quality_probability_y: 5490,
quality_probability_x: 5490)
Coordinates: (12/63)
* conditions_geometry_angle (conditions_geometry_angle) <U7 56B ...
* conditions_geometry_band (conditions_geometry_band) <U3 156B ...
* conditions_geometry_detector (conditions_geometry_detector) int64 32B ...
* conditions_geometry_x (conditions_geometry_x) int64 184B ...
* conditions_geometry_y (conditions_geometry_y) int64 184B ...
* conditions_mask_detector_footprint_r10m_x (conditions_mask_detector_footprint_r10m_x) int64 88kB ...
... ...
* quality_mask_r20m_y (quality_mask_r20m_y) int64 44kB ...
* quality_mask_r60m_x (quality_mask_r60m_x) int64 15kB ...
* quality_mask_r60m_y (quality_mask_r60m_y) int64 15kB ...
quality_probability_band int64 8B ...
* quality_probability_x (quality_probability_x) int64 44kB ...
* quality_probability_y (quality_probability_y) int64 44kB ...
Data variables: (12/86)
conditions_geometry_mean_sun_angles (conditions_geometry_angle) float64 16B dask.array<chunksize=(2,), meta=np.ndarray>
conditions_geometry_mean_viewing_incidence_angles (conditions_geometry_band, conditions_geometry_angle) float64 208B dask.array<chunksize=(13, 2), meta=np.ndarray>
conditions_geometry_sun_angles (conditions_geometry_angle, conditions_geometry_y, conditions_geometry_x) float64 8kB dask.array<chunksize=(2, 23, 23), meta=np.ndarray>
conditions_geometry_viewing_incidence_angles (conditions_geometry_band, conditions_geometry_detector, conditions_geometry_angle, conditions_geometry_y, conditions_geometry_x) float64 440kB dask.array<chunksize=(7, 4, 2, 23, 23), meta=np.ndarray>
conditions_mask_detector_footprint_r10m_b02 (conditions_mask_detector_footprint_r10m_y, conditions_mask_detector_footprint_r10m_x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray>
conditions_mask_detector_footprint_r10m_b03 (conditions_mask_detector_footprint_r10m_y, conditions_mask_detector_footprint_r10m_x) uint8 121MB dask.array<chunksize=(1830, 1830), meta=np.ndarray>
... ...
quality_mask_r20m_b8a (quality_mask_r20m_y, quality_mask_r20m_x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray>
quality_mask_r60m_b01 (quality_mask_r60m_y, quality_mask_r60m_x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray>
quality_mask_r60m_b09 (quality_mask_r60m_y, quality_mask_r60m_x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray>
quality_mask_r60m_b10 (quality_mask_r60m_y, quality_mask_r60m_x) uint8 3MB dask.array<chunksize=(305, 305), meta=np.ndarray>
quality_probability_cld (quality_probability_y, quality_probability_x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray>
quality_probability_snw (quality_probability_y, quality_probability_x) uint8 30MB dask.array<chunksize=(915, 915), meta=np.ndarray>
Attributes: (2)- conditions_geometry_angle: 2
- conditions_geometry_band: 13
- conditions_geometry_y: 23
- conditions_geometry_x: 23
- conditions_geometry_detector: 4
- conditions_mask_detector_footprint_r10m_y: 10980
- conditions_mask_detector_footprint_r10m_x: 10980
- conditions_mask_detector_footprint_r20m_y: 5490
- conditions_mask_detector_footprint_r20m_x: 5490
- conditions_mask_detector_footprint_r60m_y: 1830
- conditions_mask_detector_footprint_r60m_x: 1830
- conditions_mask_l1c_classification_y: 1830
- conditions_mask_l1c_classification_x: 1830
- conditions_mask_l2a_classification_r20m_y: 5490
- conditions_mask_l2a_classification_r20m_x: 5490
- conditions_mask_l2a_classification_r60m_y: 1830
- conditions_mask_l2a_classification_r60m_x: 1830
- conditions_meteorology_cams_latitude: 9
- conditions_meteorology_cams_longitude: 9
- conditions_meteorology_ecmwf_latitude: 9
- conditions_meteorology_ecmwf_longitude: 9
- measurements_r10m_y: 10980
- measurements_r10m_x: 10980
- measurements_r20m_y: 5490
- measurements_r20m_x: 5490
- measurements_r60m_y: 1830
- measurements_r60m_x: 1830
- quality_atmosphere_r10m_y: 10980
- quality_atmosphere_r10m_x: 10980
- quality_atmosphere_r20m_y: 5490
- quality_atmosphere_r20m_x: 5490
- quality_atmosphere_r60m_y: 1830
- quality_atmosphere_r60m_x: 1830
- quality_l2a_quicklook_r10m_band: 3
- quality_l2a_quicklook_r10m_y: 10980
- quality_l2a_quicklook_r10m_x: 10980
- quality_l2a_quicklook_r20m_band: 3
- quality_l2a_quicklook_r20m_y: 5490
- quality_l2a_quicklook_r20m_x: 5490
- quality_l2a_quicklook_r60m_band: 3
- quality_l2a_quicklook_r60m_y: 1830
- quality_l2a_quicklook_r60m_x: 1830
- quality_mask_r10m_y: 10980
- quality_mask_r10m_x: 10980
- quality_mask_r20m_y: 5490
- quality_mask_r20m_x: 5490
- quality_mask_r60m_y: 1830
- quality_mask_r60m_x: 1830
- quality_probability_y: 5490
- quality_probability_x: 5490
- conditions_geometry_angle(conditions_geometry_angle)<U7'zenith' 'azimuth'
array(['zenith', 'azimuth'], dtype='<U7')
- conditions_geometry_band(conditions_geometry_band)<U3'b01' 'b02' 'b03' ... 'b11' 'b12'
array(['b01', 'b02', 'b03', 'b04', 'b05', 'b06', 'b07', 'b08', 'b8a', 'b09', 'b10', 'b11', 'b12'], dtype='<U3') - conditions_geometry_detector(conditions_geometry_detector)int641 2 3 4
array([1, 2, 3, 4])
- conditions_geometry_x(conditions_geometry_x)int64300000 305000 ... 405000 410000
array([300000, 305000, 310000, 315000, 320000, 325000, 330000, 335000, 340000, 345000, 350000, 355000, 360000, 365000, 370000, 375000, 380000, 385000, 390000, 395000, 400000, 405000, 410000]) - conditions_geometry_y(conditions_geometry_y)int645000040 4995040 ... 4895040 4890040
array([5000040, 4995040, 4990040, 4985040, 4980040, 4975040, 4970040, 4965040, 4960040, 4955040, 4950040, 4945040, 4940040, 4935040, 4930040, 4925040, 4920040, 4915040, 4910040, 4905040, 4900040, 4895040, 4890040]) - conditions_mask_detector_footprint_r10m_x(conditions_mask_detector_footprint_r10m_x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- conditions_mask_detector_footprint_r10m_y(conditions_mask_detector_footprint_r10m_y)int645000035 5000025 ... 4890255 4890245
array([5000035, 5000025, 5000015, ..., 4890265, 4890255, 4890245])
- conditions_mask_detector_footprint_r20m_x(conditions_mask_detector_footprint_r20m_x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- conditions_mask_detector_footprint_r20m_y(conditions_mask_detector_footprint_r20m_y)int645000030 5000010 ... 4890270 4890250
array([5000030, 5000010, 4999990, ..., 4890290, 4890270, 4890250])
- conditions_mask_detector_footprint_r60m_x(conditions_mask_detector_footprint_r60m_x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- conditions_mask_detector_footprint_r60m_y(conditions_mask_detector_footprint_r60m_y)int645000010 4999950 ... 4890330 4890270
array([5000010, 4999950, 4999890, ..., 4890390, 4890330, 4890270])
- conditions_mask_l1c_classification_x(conditions_mask_l1c_classification_x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- conditions_mask_l1c_classification_y(conditions_mask_l1c_classification_y)int645000010 4999950 ... 4890330 4890270
array([5000010, 4999950, 4999890, ..., 4890390, 4890330, 4890270])
- conditions_mask_l2a_classification_r20m_x(conditions_mask_l2a_classification_r20m_x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- conditions_mask_l2a_classification_r20m_y(conditions_mask_l2a_classification_r20m_y)int645000030 5000010 ... 4890270 4890250
array([5000030, 5000010, 4999990, ..., 4890290, 4890270, 4890250])
- conditions_mask_l2a_classification_r60m_x(conditions_mask_l2a_classification_r60m_x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- conditions_mask_l2a_classification_r60m_y(conditions_mask_l2a_classification_r60m_y)int645000010 4999950 ... 4890330 4890270
array([5000010, 4999950, 4999890, ..., 4890390, 4890330, 4890270])
- conditions_meteorology_cams_isobaricInhPa()float64...
- long_name :
- pressure
- positive :
- down
- standard_name :
- air_pressure
- stored_direction :
- decreasing
- units :
- hPa
[1 values with dtype=float64]
- conditions_meteorology_cams_latitude(conditions_meteorology_cams_latitude)float6445.13 45.0 44.88 ... 44.28 44.16
- long_name :
- latitude
- standard_name :
- latitude
- stored_direction :
- decreasing
- units :
- degrees_north
array([45.126, 45.005, 44.884, 44.763, 44.642, 44.521, 44.4 , 44.279, 44.16 ])
- conditions_meteorology_cams_longitude(conditions_meteorology_cams_longitude)float646.457 6.634 6.811 ... 7.695 7.872
- long_name :
- longitude
- standard_name :
- longitude
- units :
- degrees_east
array([6.457 , 6.633875, 6.81075 , 6.987625, 7.1645 , 7.341375, 7.51825 , 7.695125, 7.872 ]) - conditions_meteorology_cams_number()int64...
- long_name :
- ensemble member numerical id
- standard_name :
- realization
- units :
- 1
[1 values with dtype=int64]
- conditions_meteorology_cams_step()int64...
- long_name :
- time since forecast_reference_time
- standard_name :
- forecast_period
- units :
- nanoseconds
[1 values with dtype=int64]
- conditions_meteorology_cams_surface()float64...
- long_name :
- original GRIB coordinate for key: level(surface)
- units :
- 1
[1 values with dtype=float64]
- conditions_meteorology_cams_time()datetime64[ns]...
- long_name :
- initial time of forecast
- standard_name :
- forecast_reference_time
[1 values with dtype=datetime64[ns]]
- conditions_meteorology_cams_valid_time()datetime64[ns]...
- long_name :
- time
- standard_name :
- time
[1 values with dtype=datetime64[ns]]
- conditions_meteorology_ecmwf_isobaricInhPa()float64...
- long_name :
- pressure
- positive :
- down
- standard_name :
- air_pressure
- stored_direction :
- decreasing
- units :
- hPa
[1 values with dtype=float64]
- conditions_meteorology_ecmwf_latitude(conditions_meteorology_ecmwf_latitude)float6445.13 45.0 44.88 ... 44.28 44.16
- long_name :
- latitude
- standard_name :
- latitude
- stored_direction :
- decreasing
- units :
- degrees_north
array([45.126, 45.005, 44.884, 44.763, 44.642, 44.521, 44.4 , 44.279, 44.16 ])
- conditions_meteorology_ecmwf_longitude(conditions_meteorology_ecmwf_longitude)float646.457 6.634 6.811 ... 7.695 7.872
- long_name :
- longitude
- standard_name :
- longitude
- units :
- degrees_east
array([6.457 , 6.633875, 6.81075 , 6.987625, 7.1645 , 7.341375, 7.51825 , 7.695125, 7.872 ]) - conditions_meteorology_ecmwf_number()int64...
- long_name :
- ensemble member numerical id
- standard_name :
- realization
- units :
- 1
[1 values with dtype=int64]
- conditions_meteorology_ecmwf_step()int64...
- long_name :
- time since forecast_reference_time
- standard_name :
- forecast_period
- units :
- nanoseconds
[1 values with dtype=int64]
- conditions_meteorology_ecmwf_surface()float64...
- long_name :
- original GRIB coordinate for key: level(surface)
- units :
- 1
[1 values with dtype=float64]
- conditions_meteorology_ecmwf_time()datetime64[ns]...
- long_name :
- initial time of forecast
- standard_name :
- forecast_reference_time
[1 values with dtype=datetime64[ns]]
- conditions_meteorology_ecmwf_valid_time()datetime64[ns]...
- long_name :
- time
- standard_name :
- time
[1 values with dtype=datetime64[ns]]
- measurements_r10m_x(measurements_r10m_x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- measurements_r10m_y(measurements_r10m_y)int645000035 5000025 ... 4890255 4890245
array([5000035, 5000025, 5000015, ..., 4890265, 4890255, 4890245])
- measurements_r20m_x(measurements_r20m_x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- measurements_r20m_y(measurements_r20m_y)int645000030 5000010 ... 4890270 4890250
array([5000030, 5000010, 4999990, ..., 4890290, 4890270, 4890250])
- measurements_r60m_x(measurements_r60m_x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- measurements_r60m_y(measurements_r60m_y)int645000010 4999950 ... 4890330 4890270
array([5000010, 4999950, 4999890, ..., 4890390, 4890330, 4890270])
- quality_atmosphere_r10m_x(quality_atmosphere_r10m_x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- quality_atmosphere_r10m_y(quality_atmosphere_r10m_y)int645000035 5000025 ... 4890255 4890245
array([5000035, 5000025, 5000015, ..., 4890265, 4890255, 4890245])
- quality_atmosphere_r20m_x(quality_atmosphere_r20m_x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- quality_atmosphere_r20m_y(quality_atmosphere_r20m_y)int645000030 5000010 ... 4890270 4890250
array([5000030, 5000010, 4999990, ..., 4890290, 4890270, 4890250])
- quality_atmosphere_r60m_x(quality_atmosphere_r60m_x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- quality_atmosphere_r60m_y(quality_atmosphere_r60m_y)int645000010 4999950 ... 4890330 4890270
array([5000010, 4999950, 4999890, ..., 4890390, 4890330, 4890270])
- quality_l2a_quicklook_r10m_band(quality_l2a_quicklook_r10m_band)int641 2 3
array([1, 2, 3])
- quality_l2a_quicklook_r10m_x(quality_l2a_quicklook_r10m_x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- quality_l2a_quicklook_r10m_y(quality_l2a_quicklook_r10m_y)int645000035 5000025 ... 4890255 4890245
array([5000035, 5000025, 5000015, ..., 4890265, 4890255, 4890245])
- quality_l2a_quicklook_r20m_band(quality_l2a_quicklook_r20m_band)int641 2 3
array([1, 2, 3])
- quality_l2a_quicklook_r20m_x(quality_l2a_quicklook_r20m_x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- quality_l2a_quicklook_r20m_y(quality_l2a_quicklook_r20m_y)int645000030 5000010 ... 4890270 4890250
array([5000030, 5000010, 4999990, ..., 4890290, 4890270, 4890250])
- quality_l2a_quicklook_r60m_band(quality_l2a_quicklook_r60m_band)int641 2 3
array([1, 2, 3])
- quality_l2a_quicklook_r60m_x(quality_l2a_quicklook_r60m_x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- quality_l2a_quicklook_r60m_y(quality_l2a_quicklook_r60m_y)int645000010 4999950 ... 4890330 4890270
array([5000010, 4999950, 4999890, ..., 4890390, 4890330, 4890270])
- quality_mask_r10m_x(quality_mask_r10m_x)int64300005 300015 ... 409785 409795
array([300005, 300015, 300025, ..., 409775, 409785, 409795])
- quality_mask_r10m_y(quality_mask_r10m_y)int645000035 5000025 ... 4890255 4890245
array([5000035, 5000025, 5000015, ..., 4890265, 4890255, 4890245])
- quality_mask_r20m_x(quality_mask_r20m_x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- quality_mask_r20m_y(quality_mask_r20m_y)int645000030 5000010 ... 4890270 4890250
array([5000030, 5000010, 4999990, ..., 4890290, 4890270, 4890250])
- quality_mask_r60m_x(quality_mask_r60m_x)int64300030 300090 ... 409710 409770
array([300030, 300090, 300150, ..., 409650, 409710, 409770])
- quality_mask_r60m_y(quality_mask_r60m_y)int645000010 4999950 ... 4890330 4890270
array([5000010, 4999950, 4999890, ..., 4890390, 4890330, 4890270])
- quality_probability_band()int64...
[1 values with dtype=int64]
- quality_probability_x(quality_probability_x)int64300010 300030 ... 409770 409790
array([300010, 300030, 300050, ..., 409750, 409770, 409790])
- quality_probability_y(quality_probability_y)int645000030 5000010 ... 4890270 4890250
array([5000030, 5000010, 4999990, ..., 4890290, 4890270, 4890250])
- conditions_geometry_mean_sun_angles(conditions_geometry_angle)float64dask.array<chunksize=(2,), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['angle'], 'dimensions': ['angle']}
- unit :
- deg
Array Chunk Bytes 16 B 16 B Shape (2,) (2,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - conditions_geometry_mean_viewing_incidence_angles(conditions_geometry_band, conditions_geometry_angle)float64dask.array<chunksize=(13, 2), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['angle', 'band'], 'dimensions': ['band', 'angle']}
- unit :
- deg
Array Chunk Bytes 208 B 208 B Shape (13, 2) (13, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - conditions_geometry_sun_angles(conditions_geometry_angle, conditions_geometry_y, conditions_geometry_x)float64dask.array<chunksize=(2, 23, 23), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['angle', 'y', 'x'], 'dimensions': ['angle', 'y', 'x']}
Array Chunk Bytes 8.27 kiB 8.27 kiB Shape (2, 23, 23) (2, 23, 23) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - conditions_geometry_viewing_incidence_angles(conditions_geometry_band, conditions_geometry_detector, conditions_geometry_angle, conditions_geometry_y, conditions_geometry_x)float64dask.array<chunksize=(7, 4, 2, 23, 23), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['angle', 'y', 'x', 'detector', 'band'], 'dimensions': ['band', 'detector', 'angle', 'y', 'x']}
Array Chunk Bytes 429.81 kiB 231.44 kiB Shape (13, 4, 2, 23, 23) (7, 4, 2, 23, 23) Dask graph 2 chunks in 2 graph layers Data type float64 numpy.ndarray - conditions_mask_detector_footprint_r10m_b02(conditions_mask_detector_footprint_r10m_y, conditions_mask_detector_footprint_r10m_x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r10m_b03(conditions_mask_detector_footprint_r10m_y, conditions_mask_detector_footprint_r10m_x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r10m_b04(conditions_mask_detector_footprint_r10m_y, conditions_mask_detector_footprint_r10m_x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r10m_b08(conditions_mask_detector_footprint_r10m_y, conditions_mask_detector_footprint_r10m_x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r20m_b05(conditions_mask_detector_footprint_r20m_y, conditions_mask_detector_footprint_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r20m_b06(conditions_mask_detector_footprint_r20m_y, conditions_mask_detector_footprint_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r20m_b07(conditions_mask_detector_footprint_r20m_y, conditions_mask_detector_footprint_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r20m_b11(conditions_mask_detector_footprint_r20m_y, conditions_mask_detector_footprint_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r20m_b12(conditions_mask_detector_footprint_r20m_y, conditions_mask_detector_footprint_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r20m_b8a(conditions_mask_detector_footprint_r20m_y, conditions_mask_detector_footprint_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r60m_b01(conditions_mask_detector_footprint_r60m_y, conditions_mask_detector_footprint_r60m_x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r60m_b09(conditions_mask_detector_footprint_r60m_y, conditions_mask_detector_footprint_r60m_x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_detector_footprint_r60m_b10(conditions_mask_detector_footprint_r60m_y, conditions_mask_detector_footprint_r60m_x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'long_name': 'detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12'}
- dtype :
- <u1
- long_name :
- detector footprint mask provided in the final reference frame (ground geometry). 0 = no detector, 1-12 = detector 1-12
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_l1c_classification_b00(conditions_mask_l1c_classification_y, conditions_mask_l1c_classification_x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4], 'flag_meanings': ['OPAQUE', 'CIRRUS', 'SNOW_ICE'], 'long_name': 'cloud classification mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4]
- flag_meanings :
- ['OPAQUE', 'CIRRUS', 'SNOW_ICE']
- long_name :
- cloud classification mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_l2a_classification_r20m_scl(conditions_mask_l2a_classification_r20m_y, conditions_mask_l2a_classification_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_mask_l2a_classification_r60m_scl(conditions_mask_l2a_classification_r60m_y, conditions_mask_l2a_classification_r60m_x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - conditions_meteorology_cams_aod1240(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod1240
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 1240nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210216
- GRIB_shortName :
- aod1240
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 1240nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 1240nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_aod469(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod469
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 469nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210213
- GRIB_shortName :
- aod469
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 469nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 469nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_aod550(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210207
- GRIB_shortName :
- aod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_aod670(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod670
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 670nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210214
- GRIB_shortName :
- aod670
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 670nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 670nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_aod865(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- aod865
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total Aerosol Optical Depth at 865nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210215
- GRIB_shortName :
- aod865
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total Aerosol Optical Depth at 865nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Total Aerosol Optical Depth at 865nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_bcaod550(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- bcaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Black Carbon Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210211
- GRIB_shortName :
- bcaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Black Carbon Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Black Carbon Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_duaod550(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- duaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Dust Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210209
- GRIB_shortName :
- duaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- isobaricInhPa
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Dust Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Dust Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_omaod550(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- omaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Organic Matter Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210210
- GRIB_shortName :
- omaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Organic Matter Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Organic Matter Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_ssaod550(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- ssaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Sea Salt Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210208
- GRIB_shortName :
- ssaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Sea Salt Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Sea Salt Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_suaod550(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- suaod550
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Sulphate Aerosol Optical Depth at 550nm
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 210212
- GRIB_shortName :
- suaod550
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- ~
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Sulphate Aerosol Optical Depth at 550nm', 'standard_name': 'unknown', 'units': '~'}
- long_name :
- Sulphate Aerosol Optical Depth at 550nm
- standard_name :
- unknown
- units :
- ~
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_cams_z(conditions_meteorology_cams_latitude, conditions_meteorology_cams_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- geopotential
- GRIB_cfVarName :
- z
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Geopotential
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 129
- GRIB_shortName :
- z
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- m**2 s**-2
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Geopotential', 'standard_name': 'geopotential', 'units': 'm**2 s**-2'}
- long_name :
- Geopotential
- standard_name :
- geopotential
- units :
- m**2 s**-2
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_ecmwf_msl(conditions_meteorology_ecmwf_latitude, conditions_meteorology_ecmwf_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- air_pressure_at_mean_sea_level
- GRIB_cfVarName :
- msl
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Mean sea level pressure
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 151
- GRIB_shortName :
- msl
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- Pa
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Mean sea level pressure', 'standard_name': 'air_pressure_at_mean_sea_level', 'units': 'Pa'}
- long_name :
- Mean sea level pressure
- standard_name :
- air_pressure_at_mean_sea_level
- units :
- Pa
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_ecmwf_r(conditions_meteorology_ecmwf_latitude, conditions_meteorology_ecmwf_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- relative_humidity
- GRIB_cfVarName :
- r
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Relative humidity
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 157
- GRIB_shortName :
- r
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- isobaricInhPa
- GRIB_units :
- %
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Relative humidity', 'standard_name': 'relative_humidity', 'units': '%'}
- long_name :
- Relative humidity
- standard_name :
- relative_humidity
- units :
- %
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_ecmwf_tco3(conditions_meteorology_ecmwf_latitude, conditions_meteorology_ecmwf_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- atmosphere_mass_content_of_ozone
- GRIB_cfVarName :
- tco3
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total column ozone
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 206
- GRIB_shortName :
- tco3
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- kg m**-2
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total column ozone', 'standard_name': 'atmosphere_mass_content_of_ozone', 'units': 'kg m**-2'}
- long_name :
- Total column ozone
- standard_name :
- atmosphere_mass_content_of_ozone
- units :
- kg m**-2
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_ecmwf_tcwv(conditions_meteorology_ecmwf_latitude, conditions_meteorology_ecmwf_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- lwe_thickness_of_atmosphere_mass_content_of_water_vapor
- GRIB_cfVarName :
- tcwv
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- Total column vertically-integrated water vapour
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 137
- GRIB_shortName :
- tcwv
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- kg m**-2
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': 'Total column vertically-integrated water vapour', 'standard_name': 'lwe_thickness_of_atmosphere_mass_content_of_water_vapor', 'units': 'kg m**-2'}
- long_name :
- Total column vertically-integrated water vapour
- standard_name :
- lwe_thickness_of_atmosphere_mass_content_of_water_vapor
- units :
- kg m**-2
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_ecmwf_u10(conditions_meteorology_ecmwf_latitude, conditions_meteorology_ecmwf_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- u10
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- 10 metre U wind component
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 165
- GRIB_shortName :
- 10u
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- m s**-1
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': '10 metre U wind component', 'standard_name': 'unknown', 'units': 'm s**-1'}
- long_name :
- 10 metre U wind component
- standard_name :
- unknown
- units :
- m s**-1
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - conditions_meteorology_ecmwf_v10(conditions_meteorology_ecmwf_latitude, conditions_meteorology_ecmwf_longitude)float32dask.array<chunksize=(9, 9), meta=np.ndarray>
- GRIB_NV :
- 0
- GRIB_Nx :
- 9
- GRIB_Ny :
- 9
- GRIB_cfName :
- unknown
- GRIB_cfVarName :
- v10
- GRIB_dataType :
- fc
- GRIB_gridDefinitionDescription :
- Latitude/Longitude Grid
- GRIB_gridType :
- regular_ll
- GRIB_iDirectionIncrementInDegrees :
- 0.177
- GRIB_iScansNegatively :
- 0
- GRIB_jDirectionIncrementInDegrees :
- 0.121
- GRIB_jPointsAreConsecutive :
- 0
- GRIB_jScansPositively :
- 0
- GRIB_latitudeOfFirstGridPointInDegrees :
- 45.126
- GRIB_latitudeOfLastGridPointInDegrees :
- 44.16
- GRIB_longitudeOfFirstGridPointInDegrees :
- 6.457
- GRIB_longitudeOfLastGridPointInDegrees :
- 7.872
- GRIB_missingValue :
- 3.4028234663852886e+38
- GRIB_name :
- 10 metre V wind component
- GRIB_numberOfPoints :
- 81
- GRIB_paramId :
- 166
- GRIB_shortName :
- 10v
- GRIB_stepType :
- instant
- GRIB_stepUnits :
- 0
- GRIB_totalNumber :
- 0
- GRIB_typeOfLevel :
- surface
- GRIB_units :
- m s**-1
- _eopf_attrs :
- {'coordinates': ['number', 'time', 'step', 'surface', 'latitude', 'longitude', 'valid_time', 'isobaricInhPa'], 'dimensions': ['latitude', 'longitude'], 'long_name': '10 metre V wind component', 'standard_name': 'unknown', 'units': 'm s**-1'}
- long_name :
- 10 metre V wind component
- standard_name :
- unknown
- units :
- m s**-1
Array Chunk Bytes 324 B 324 B Shape (9, 9) (9, 9) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - measurements_r10m_b02(measurements_r10m_y, measurements_r10m_x)float64dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b02 492.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b02 492.3 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 919.80 MiB 25.55 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r10m_b03(measurements_r10m_y, measurements_r10m_x)float64dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b03 559.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b03 559.0 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 919.80 MiB 25.55 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r10m_b04(measurements_r10m_y, measurements_r10m_x)float64dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b04 665.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b04 665.0 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 919.80 MiB 25.55 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r10m_b08(measurements_r10m_y, measurements_r10m_x)float64dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b08 833.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b08 833.0 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 919.80 MiB 25.55 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b01(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b01 442.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b01 442.3 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b02(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b02 492.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b02 492.3 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b03(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b03 559.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b03 559.0 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b04(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b04 665.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b04 665.0 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b05(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b05 703.8 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b05 703.8 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b06(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b06 739.1 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b06 739.1 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b07(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b07 779.7 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b07 779.7 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b11(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b11 1610.4 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b11 1610.4 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b12(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b12 2185.7 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b12 2185.7 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r20m_b8a(measurements_r20m_y, measurements_r20m_x)float64dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b8a 864.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b8a 864.0 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b01(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b01 442.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b01 442.3 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b02(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b02 492.3 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b02 492.3 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b03(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b03 559.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b03 559.0 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b04(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b04 665.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b04 665.0 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b05(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b05 703.8 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b05 703.8 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b06(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b06 739.1 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b06 739.1 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b07(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b07 779.7 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b07 779.7 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b09(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b09 943.2 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b09 943.2 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b11(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b11 1610.4 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b11 1610.4 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b12(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b12 2185.7 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b12 2185.7 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - measurements_r60m_b8a(measurements_r60m_y, measurements_r60m_x)float64dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'add_offset': -0.1, 'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u2', 'fill_value': 0, 'long_name': 'BOA reflectance from MSI acquisition at spectral band b8a 864.0 nm', 'scale_factor': 0.0001, 'units': 'digital_counts'}
- dtype :
- <u2
- fill_value :
- 0
- long_name :
- BOA reflectance from MSI acquisition at spectral band b8a 864.0 nm
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
- units :
- digital_counts
- valid_max :
- 65535
- valid_min :
- 1
Array Chunk Bytes 25.55 MiB 726.76 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type float64 numpy.ndarray - quality_atmosphere_r10m_aot(quality_atmosphere_r10m_y, quality_atmosphere_r10m_x)uint16dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray - quality_atmosphere_r10m_wvp(quality_atmosphere_r10m_y, quality_atmosphere_r10m_x)uint16dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 229.95 MiB 6.39 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray - quality_atmosphere_r20m_aot(quality_atmosphere_r20m_y, quality_atmosphere_r20m_x)uint16dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 57.49 MiB 1.60 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray - quality_atmosphere_r20m_wvp(quality_atmosphere_r20m_y, quality_atmosphere_r20m_x)uint16dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 57.49 MiB 1.60 MiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray - quality_atmosphere_r60m_aot(quality_atmosphere_r60m_y, quality_atmosphere_r60m_x)uint16dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 6.39 MiB 181.69 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray - quality_atmosphere_r60m_wvp(quality_atmosphere_r60m_y, quality_atmosphere_r60m_x)uint16dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 6.39 MiB 181.69 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint16 numpy.ndarray - quality_l2a_quicklook_r10m_tci(quality_l2a_quicklook_r10m_band, quality_l2a_quicklook_r10m_y, quality_l2a_quicklook_r10m_x)uint8dask.array<chunksize=(1, 1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 344.93 MiB 3.19 MiB Shape (3, 10980, 10980) (1, 1830, 1830) Dask graph 108 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_l2a_quicklook_r20m_tci(quality_l2a_quicklook_r20m_band, quality_l2a_quicklook_r20m_y, quality_l2a_quicklook_r20m_x)uint8dask.array<chunksize=(1, 915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 86.23 MiB 817.60 kiB Shape (3, 5490, 5490) (1, 915, 915) Dask graph 108 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_l2a_quicklook_r60m_tci(quality_l2a_quicklook_r60m_band, quality_l2a_quicklook_r60m_y, quality_l2a_quicklook_r60m_x)uint8dask.array<chunksize=(1, 305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['band', 'y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 9.58 MiB 90.84 kiB Shape (3, 1830, 1830) (1, 305, 305) Dask graph 108 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r10m_b02(quality_mask_r10m_y, quality_mask_r10m_x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r10m_b03(quality_mask_r10m_y, quality_mask_r10m_x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r10m_b04(quality_mask_r10m_y, quality_mask_r10m_x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r10m_b08(quality_mask_r10m_y, quality_mask_r10m_x)uint8dask.array<chunksize=(1830, 1830), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [10980, 10980]
- proj:transform :
- [10.0, 0.0, 300000.0, 0.0, -10.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 114.98 MiB 3.19 MiB Shape (10980, 10980) (1830, 1830) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r20m_b05(quality_mask_r20m_y, quality_mask_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r20m_b06(quality_mask_r20m_y, quality_mask_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r20m_b07(quality_mask_r20m_y, quality_mask_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r20m_b11(quality_mask_r20m_y, quality_mask_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r20m_b12(quality_mask_r20m_y, quality_mask_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r20m_b8a(quality_mask_r20m_y, quality_mask_r20m_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r60m_b01(quality_mask_r60m_y, quality_mask_r60m_x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r60m_b09(quality_mask_r60m_y, quality_mask_r60m_x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_mask_r60m_b10(quality_mask_r60m_y, quality_mask_r60m_x)uint8dask.array<chunksize=(305, 305), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['x', 'y'], 'dimensions': ['y', 'x'], 'dtype': '<u1', 'flag_masks': [1, 2, 4, 8, 16, 32, 64, 128], 'flag_meanings': ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A'], 'long_name': 'quality mask provided in the final reference frame (ground geometry)'}
- dtype :
- <u1
- flag_masks :
- [1, 2, 4, 8, 16, 32, 64, 128]
- flag_meanings :
- ['ANC_LOST', 'ANC_DEG', 'MSI_LOST', 'MSI_DEG', 'QT_DEFECTIVE_PIXELS', 'QT_NODATA_PIXELS', 'QT_PARTIALLY_CORRECTED_PIXELS', 'QT_SATURATED_PIXELS_L1A']
- long_name :
- quality mask provided in the final reference frame (ground geometry)
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [1830, 1830]
- proj:transform :
- [60.0, 0.0, 300000.0, 0.0, -60.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 3.19 MiB 90.84 kiB Shape (1830, 1830) (305, 305) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_probability_cld(quality_probability_y, quality_probability_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray - quality_probability_snw(quality_probability_y, quality_probability_x)uint8dask.array<chunksize=(915, 915), meta=np.ndarray>
- _eopf_attrs :
- {'coordinates': ['band', 'x', 'y'], 'dimensions': ['y', 'x']}
- proj:bbox :
- [300000.0, 4890240.0, 409800.0, 5000040.0]
- proj:epsg :
- 32632
- proj:shape :
- [5490, 5490]
- proj:transform :
- [20.0, 0.0, 300000.0, 0.0, -20.0, 5000040.0, 0.0, 0.0, 1.0]
- proj:wkt2 :
- PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]
Array Chunk Bytes 28.74 MiB 817.60 kiB Shape (5490, 5490) (915, 915) Dask graph 36 chunks in 2 graph layers Data type uint8 numpy.ndarray
- conditions_geometry_anglePandasIndex
PandasIndex(Index(['zenith', 'azimuth'], dtype='object', name='conditions_geometry_angle'))
- conditions_geometry_bandPandasIndex
PandasIndex(Index(['b01', 'b02', 'b03', 'b04', 'b05', 'b06', 'b07', 'b08', 'b8a', 'b09', 'b10', 'b11', 'b12'], dtype='object', name='conditions_geometry_band')) - conditions_geometry_detectorPandasIndex
PandasIndex(Index([1, 2, 3, 4], dtype='int64', name='conditions_geometry_detector'))
- conditions_geometry_xPandasIndex
PandasIndex(Index([300000, 305000, 310000, 315000, 320000, 325000, 330000, 335000, 340000, 345000, 350000, 355000, 360000, 365000, 370000, 375000, 380000, 385000, 390000, 395000, 400000, 405000, 410000], dtype='int64', name='conditions_geometry_x')) - conditions_geometry_yPandasIndex
PandasIndex(Index([5000040, 4995040, 4990040, 4985040, 4980040, 4975040, 4970040, 4965040, 4960040, 4955040, 4950040, 4945040, 4940040, 4935040, 4930040, 4925040, 4920040, 4915040, 4910040, 4905040, 4900040, 4895040, 4890040], dtype='int64', name='conditions_geometry_y')) - conditions_mask_detector_footprint_r10m_xPandasIndex
PandasIndex(Index([300005, 300015, 300025, 300035, 300045, 300055, 300065, 300075, 300085, 300095, ... 409705, 409715, 409725, 409735, 409745, 409755, 409765, 409775, 409785, 409795], dtype='int64', name='conditions_mask_detector_footprint_r10m_x', length=10980)) - conditions_mask_detector_footprint_r10m_yPandasIndex
PandasIndex(Index([5000035, 5000025, 5000015, 5000005, 4999995, 4999985, 4999975, 4999965, 4999955, 4999945, ... 4890335, 4890325, 4890315, 4890305, 4890295, 4890285, 4890275, 4890265, 4890255, 4890245], dtype='int64', name='conditions_mask_detector_footprint_r10m_y', length=10980)) - conditions_mask_detector_footprint_r20m_xPandasIndex
PandasIndex(Index([300010, 300030, 300050, 300070, 300090, 300110, 300130, 300150, 300170, 300190, ... 409610, 409630, 409650, 409670, 409690, 409710, 409730, 409750, 409770, 409790], dtype='int64', name='conditions_mask_detector_footprint_r20m_x', length=5490)) - conditions_mask_detector_footprint_r20m_yPandasIndex
PandasIndex(Index([5000030, 5000010, 4999990, 4999970, 4999950, 4999930, 4999910, 4999890, 4999870, 4999850, ... 4890430, 4890410, 4890390, 4890370, 4890350, 4890330, 4890310, 4890290, 4890270, 4890250], dtype='int64', name='conditions_mask_detector_footprint_r20m_y', length=5490)) - conditions_mask_detector_footprint_r60m_xPandasIndex
PandasIndex(Index([300030, 300090, 300150, 300210, 300270, 300330, 300390, 300450, 300510, 300570, ... 409230, 409290, 409350, 409410, 409470, 409530, 409590, 409650, 409710, 409770], dtype='int64', name='conditions_mask_detector_footprint_r60m_x', length=1830)) - conditions_mask_detector_footprint_r60m_yPandasIndex
PandasIndex(Index([5000010, 4999950, 4999890, 4999830, 4999770, 4999710, 4999650, 4999590, 4999530, 4999470, ... 4890810, 4890750, 4890690, 4890630, 4890570, 4890510, 4890450, 4890390, 4890330, 4890270], dtype='int64', name='conditions_mask_detector_footprint_r60m_y', length=1830)) - conditions_mask_l1c_classification_xPandasIndex
PandasIndex(Index([300030, 300090, 300150, 300210, 300270, 300330, 300390, 300450, 300510, 300570, ... 409230, 409290, 409350, 409410, 409470, 409530, 409590, 409650, 409710, 409770], dtype='int64', name='conditions_mask_l1c_classification_x', length=1830)) - conditions_mask_l1c_classification_yPandasIndex
PandasIndex(Index([5000010, 4999950, 4999890, 4999830, 4999770, 4999710, 4999650, 4999590, 4999530, 4999470, ... 4890810, 4890750, 4890690, 4890630, 4890570, 4890510, 4890450, 4890390, 4890330, 4890270], dtype='int64', name='conditions_mask_l1c_classification_y', length=1830)) - conditions_mask_l2a_classification_r20m_xPandasIndex
PandasIndex(Index([300010, 300030, 300050, 300070, 300090, 300110, 300130, 300150, 300170, 300190, ... 409610, 409630, 409650, 409670, 409690, 409710, 409730, 409750, 409770, 409790], dtype='int64', name='conditions_mask_l2a_classification_r20m_x', length=5490)) - conditions_mask_l2a_classification_r20m_yPandasIndex
PandasIndex(Index([5000030, 5000010, 4999990, 4999970, 4999950, 4999930, 4999910, 4999890, 4999870, 4999850, ... 4890430, 4890410, 4890390, 4890370, 4890350, 4890330, 4890310, 4890290, 4890270, 4890250], dtype='int64', name='conditions_mask_l2a_classification_r20m_y', length=5490)) - conditions_mask_l2a_classification_r60m_xPandasIndex
PandasIndex(Index([300030, 300090, 300150, 300210, 300270, 300330, 300390, 300450, 300510, 300570, ... 409230, 409290, 409350, 409410, 409470, 409530, 409590, 409650, 409710, 409770], dtype='int64', name='conditions_mask_l2a_classification_r60m_x', length=1830)) - conditions_mask_l2a_classification_r60m_yPandasIndex
PandasIndex(Index([5000010, 4999950, 4999890, 4999830, 4999770, 4999710, 4999650, 4999590, 4999530, 4999470, ... 4890810, 4890750, 4890690, 4890630, 4890570, 4890510, 4890450, 4890390, 4890330, 4890270], dtype='int64', name='conditions_mask_l2a_classification_r60m_y', length=1830)) - conditions_meteorology_cams_latitudePandasIndex
PandasIndex(Index([ 45.126, 45.004999999999995, 44.88399999999999, 44.76299999999999, 44.64199999999999, 44.52099999999999, 44.399999999999984, 44.27899999999998, 44.16], dtype='float64', name='conditions_meteorology_cams_latitude')) - conditions_meteorology_cams_longitudePandasIndex
PandasIndex(Index([ 6.457, 6.633875, 6.81075, 6.9876249999999995, 7.164499999999999, 7.341374999999999, 7.518249999999999, 7.695124999999999, 7.872], dtype='float64', name='conditions_meteorology_cams_longitude')) - conditions_meteorology_ecmwf_latitudePandasIndex
PandasIndex(Index([ 45.126, 45.004999999999995, 44.88399999999999, 44.76299999999999, 44.64199999999999, 44.52099999999999, 44.399999999999984, 44.27899999999998, 44.16], dtype='float64', name='conditions_meteorology_ecmwf_latitude')) - conditions_meteorology_ecmwf_longitudePandasIndex
PandasIndex(Index([ 6.457, 6.633875, 6.81075, 6.9876249999999995, 7.164499999999999, 7.341374999999999, 7.518249999999999, 7.695124999999999, 7.872], dtype='float64', name='conditions_meteorology_ecmwf_longitude')) - measurements_r10m_xPandasIndex
PandasIndex(Index([300005, 300015, 300025, 300035, 300045, 300055, 300065, 300075, 300085, 300095, ... 409705, 409715, 409725, 409735, 409745, 409755, 409765, 409775, 409785, 409795], dtype='int64', name='measurements_r10m_x', length=10980)) - measurements_r10m_yPandasIndex
PandasIndex(Index([5000035, 5000025, 5000015, 5000005, 4999995, 4999985, 4999975, 4999965, 4999955, 4999945, ... 4890335, 4890325, 4890315, 4890305, 4890295, 4890285, 4890275, 4890265, 4890255, 4890245], dtype='int64', name='measurements_r10m_y', length=10980)) - measurements_r20m_xPandasIndex
PandasIndex(Index([300010, 300030, 300050, 300070, 300090, 300110, 300130, 300150, 300170, 300190, ... 409610, 409630, 409650, 409670, 409690, 409710, 409730, 409750, 409770, 409790], dtype='int64', name='measurements_r20m_x', length=5490)) - measurements_r20m_yPandasIndex
PandasIndex(Index([5000030, 5000010, 4999990, 4999970, 4999950, 4999930, 4999910, 4999890, 4999870, 4999850, ... 4890430, 4890410, 4890390, 4890370, 4890350, 4890330, 4890310, 4890290, 4890270, 4890250], dtype='int64', name='measurements_r20m_y', length=5490)) - measurements_r60m_xPandasIndex
PandasIndex(Index([300030, 300090, 300150, 300210, 300270, 300330, 300390, 300450, 300510, 300570, ... 409230, 409290, 409350, 409410, 409470, 409530, 409590, 409650, 409710, 409770], dtype='int64', name='measurements_r60m_x', length=1830)) - measurements_r60m_yPandasIndex
PandasIndex(Index([5000010, 4999950, 4999890, 4999830, 4999770, 4999710, 4999650, 4999590, 4999530, 4999470, ... 4890810, 4890750, 4890690, 4890630, 4890570, 4890510, 4890450, 4890390, 4890330, 4890270], dtype='int64', name='measurements_r60m_y', length=1830)) - quality_atmosphere_r10m_xPandasIndex
PandasIndex(Index([300005, 300015, 300025, 300035, 300045, 300055, 300065, 300075, 300085, 300095, ... 409705, 409715, 409725, 409735, 409745, 409755, 409765, 409775, 409785, 409795], dtype='int64', name='quality_atmosphere_r10m_x', length=10980)) - quality_atmosphere_r10m_yPandasIndex
PandasIndex(Index([5000035, 5000025, 5000015, 5000005, 4999995, 4999985, 4999975, 4999965, 4999955, 4999945, ... 4890335, 4890325, 4890315, 4890305, 4890295, 4890285, 4890275, 4890265, 4890255, 4890245], dtype='int64', name='quality_atmosphere_r10m_y', length=10980)) - quality_atmosphere_r20m_xPandasIndex
PandasIndex(Index([300010, 300030, 300050, 300070, 300090, 300110, 300130, 300150, 300170, 300190, ... 409610, 409630, 409650, 409670, 409690, 409710, 409730, 409750, 409770, 409790], dtype='int64', name='quality_atmosphere_r20m_x', length=5490)) - quality_atmosphere_r20m_yPandasIndex
PandasIndex(Index([5000030, 5000010, 4999990, 4999970, 4999950, 4999930, 4999910, 4999890, 4999870, 4999850, ... 4890430, 4890410, 4890390, 4890370, 4890350, 4890330, 4890310, 4890290, 4890270, 4890250], dtype='int64', name='quality_atmosphere_r20m_y', length=5490)) - quality_atmosphere_r60m_xPandasIndex
PandasIndex(Index([300030, 300090, 300150, 300210, 300270, 300330, 300390, 300450, 300510, 300570, ... 409230, 409290, 409350, 409410, 409470, 409530, 409590, 409650, 409710, 409770], dtype='int64', name='quality_atmosphere_r60m_x', length=1830)) - quality_atmosphere_r60m_yPandasIndex
PandasIndex(Index([5000010, 4999950, 4999890, 4999830, 4999770, 4999710, 4999650, 4999590, 4999530, 4999470, ... 4890810, 4890750, 4890690, 4890630, 4890570, 4890510, 4890450, 4890390, 4890330, 4890270], dtype='int64', name='quality_atmosphere_r60m_y', length=1830)) - quality_l2a_quicklook_r10m_bandPandasIndex
PandasIndex(Index([1, 2, 3], dtype='int64', name='quality_l2a_quicklook_r10m_band'))
- quality_l2a_quicklook_r10m_xPandasIndex
PandasIndex(Index([300005, 300015, 300025, 300035, 300045, 300055, 300065, 300075, 300085, 300095, ... 409705, 409715, 409725, 409735, 409745, 409755, 409765, 409775, 409785, 409795], dtype='int64', name='quality_l2a_quicklook_r10m_x', length=10980)) - quality_l2a_quicklook_r10m_yPandasIndex
PandasIndex(Index([5000035, 5000025, 5000015, 5000005, 4999995, 4999985, 4999975, 4999965, 4999955, 4999945, ... 4890335, 4890325, 4890315, 4890305, 4890295, 4890285, 4890275, 4890265, 4890255, 4890245], dtype='int64', name='quality_l2a_quicklook_r10m_y', length=10980)) - quality_l2a_quicklook_r20m_bandPandasIndex
PandasIndex(Index([1, 2, 3], dtype='int64', name='quality_l2a_quicklook_r20m_band'))
- quality_l2a_quicklook_r20m_xPandasIndex
PandasIndex(Index([300010, 300030, 300050, 300070, 300090, 300110, 300130, 300150, 300170, 300190, ... 409610, 409630, 409650, 409670, 409690, 409710, 409730, 409750, 409770, 409790], dtype='int64', name='quality_l2a_quicklook_r20m_x', length=5490)) - quality_l2a_quicklook_r20m_yPandasIndex
PandasIndex(Index([5000030, 5000010, 4999990, 4999970, 4999950, 4999930, 4999910, 4999890, 4999870, 4999850, ... 4890430, 4890410, 4890390, 4890370, 4890350, 4890330, 4890310, 4890290, 4890270, 4890250], dtype='int64', name='quality_l2a_quicklook_r20m_y', length=5490)) - quality_l2a_quicklook_r60m_bandPandasIndex
PandasIndex(Index([1, 2, 3], dtype='int64', name='quality_l2a_quicklook_r60m_band'))
- quality_l2a_quicklook_r60m_xPandasIndex
PandasIndex(Index([300030, 300090, 300150, 300210, 300270, 300330, 300390, 300450, 300510, 300570, ... 409230, 409290, 409350, 409410, 409470, 409530, 409590, 409650, 409710, 409770], dtype='int64', name='quality_l2a_quicklook_r60m_x', length=1830)) - quality_l2a_quicklook_r60m_yPandasIndex
PandasIndex(Index([5000010, 4999950, 4999890, 4999830, 4999770, 4999710, 4999650, 4999590, 4999530, 4999470, ... 4890810, 4890750, 4890690, 4890630, 4890570, 4890510, 4890450, 4890390, 4890330, 4890270], dtype='int64', name='quality_l2a_quicklook_r60m_y', length=1830)) - quality_mask_r10m_xPandasIndex
PandasIndex(Index([300005, 300015, 300025, 300035, 300045, 300055, 300065, 300075, 300085, 300095, ... 409705, 409715, 409725, 409735, 409745, 409755, 409765, 409775, 409785, 409795], dtype='int64', name='quality_mask_r10m_x', length=10980)) - quality_mask_r10m_yPandasIndex
PandasIndex(Index([5000035, 5000025, 5000015, 5000005, 4999995, 4999985, 4999975, 4999965, 4999955, 4999945, ... 4890335, 4890325, 4890315, 4890305, 4890295, 4890285, 4890275, 4890265, 4890255, 4890245], dtype='int64', name='quality_mask_r10m_y', length=10980)) - quality_mask_r20m_xPandasIndex
PandasIndex(Index([300010, 300030, 300050, 300070, 300090, 300110, 300130, 300150, 300170, 300190, ... 409610, 409630, 409650, 409670, 409690, 409710, 409730, 409750, 409770, 409790], dtype='int64', name='quality_mask_r20m_x', length=5490)) - quality_mask_r20m_yPandasIndex
PandasIndex(Index([5000030, 5000010, 4999990, 4999970, 4999950, 4999930, 4999910, 4999890, 4999870, 4999850, ... 4890430, 4890410, 4890390, 4890370, 4890350, 4890330, 4890310, 4890290, 4890270, 4890250], dtype='int64', name='quality_mask_r20m_y', length=5490)) - quality_mask_r60m_xPandasIndex
PandasIndex(Index([300030, 300090, 300150, 300210, 300270, 300330, 300390, 300450, 300510, 300570, ... 409230, 409290, 409350, 409410, 409470, 409530, 409590, 409650, 409710, 409770], dtype='int64', name='quality_mask_r60m_x', length=1830)) - quality_mask_r60m_yPandasIndex
PandasIndex(Index([5000010, 4999950, 4999890, 4999830, 4999770, 4999710, 4999650, 4999590, 4999530, 4999470, ... 4890810, 4890750, 4890690, 4890630, 4890570, 4890510, 4890450, 4890390, 4890330, 4890270], dtype='int64', name='quality_mask_r60m_y', length=1830)) - quality_probability_xPandasIndex
PandasIndex(Index([300010, 300030, 300050, 300070, 300090, 300110, 300130, 300150, 300170, 300190, ... 409610, 409630, 409650, 409670, 409690, 409710, 409730, 409750, 409770, 409790], dtype='int64', name='quality_probability_x', length=5490)) - quality_probability_yPandasIndex
PandasIndex(Index([5000030, 5000010, 4999990, 4999970, 4999950, 4999930, 4999910, 4999890, 4999870, 4999850, ... 4890430, 4890410, 4890390, 4890370, 4890350, 4890330, 4890310, 4890290, 4890270, 4890250], dtype='int64', name='quality_probability_y', length=5490))
- other_metadata :
- {'AOT_retrieval_model': 'SEN2COR_DDV', 'L0_ancillary_data_quality': '2', 'L0_ephemeris_data_quality': '3', 'NUC_table_ID': 2, 'SWIR_rearrangement_flag': 'null', 'UTM_zone_identification': 'S2B_OPER_MSI_L2A_TL_2BPS_20250430T131328_A042562_T32TLQ_N05.11', 'absolute_location_assessment_from_AOCS': '\n ', 'band_description': {'01': {'bandwidth': '20.0', 'central_wavelength': 442.3, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '7.22833', 'physical_gain': '3.95548343', 'spectral_response_step': '1', 'spectral_response_values': '0.0062411 0.01024045 0.00402983 0.00642179 0.00552753 0.0065525 0.00409887 0.006297 0.00436742 0.00233356 0.00058162 0.00202276 0.00294328 0.00485362 0.00317041 0.00237657 0.00234612 0.00440152 0.01292397 0.05001678 0.18650104 0.45441623 0.72307877 0.83999211 0.86456334 0.87472096 0.89215296 0.91090814 0.92588017 0.93924094 0.94491826 0.95078529 0.96803023 0.99939195 1 0.97548364 0.96148351 0.94986211 0.91841452 0.87989802 0.80383677 0.59752075 0.30474132 0.10798014 0.0304465 0.00885119', 'units': 'nm', 'wavelength_max': 456.0, 'wavelength_min': 411.0}, '02': {'bandwidth': '65.0', 'central_wavelength': 492.3, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.2646269', 'physical_gain': '3.81250879', 'spectral_response_step': '1', 'spectral_response_values': '0.05529541 0.12005068 0.25199051 0.4623617 0.65162379 0.77642171 0.82319091 0.83083116 0.83382106 0.837526 0.86304286 0.88226141 0.90486326 0.92043837 0.93602675 0.930533 0.92714067 0.9161479 0.90551724 0.89745515 0.90266694 0.90854264 0.92047913 0.92417935 0.91845025 0.90743244 0.89733983 0.88646415 0.87189983 0.85643973 0.84473414 0.84190734 0.85644111 0.87782724 0.90261174 0.91840544 0.94585847 0.96887192 0.99336135 0.99927899 1 0.99520325 0.98412711 0.97947473 0.97808297 0.97213439 0.96277794 0.95342234 0.93802376 0.92460144 0.90932642 0.90192251 0.89184298 0.88963556 0.89146958 0.89877911 0.91056869 0.92427362 0.93823555 0.95311791 0.97150808 0.98737003 0.99658514 0.99367959 0.98144714 0.95874415 0.89291635 0.73566218 0.52060373 0.3322804 0.19492197 0.11732617 0.07507304 0.05094154 0.03213016 0.01510217 0.00447984', 'units': 'nm', 'wavelength_max': 532.0, 'wavelength_min': 456.0}, '03': {'bandwidth': '35.0', 'central_wavelength': 559.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.2996743', 'physical_gain': '4.22866877', 'spectral_response_step': '1', 'spectral_response_values': '0.00188039 0.01545903 0.03660414 0.08100583 0.16917887 0.33278274 0.58622794 0.80916412 0.913051 0.94472284 0.94898813 0.94369132 0.92845674 0.91256938 0.90078036 0.89958598 0.90547138 0.92045355 0.94065665 0.96199681 0.98186744 0.9985841 1 0.99279888 0.97801325 0.95301174 0.9266333 0.89359131 0.86941793 0.84827 0.83908301 0.83206209 0.8291787 0.83305844 0.84630939 0.86396307 0.87268076 0.86818339 0.8554947 0.80839054 0.67650876 0.45584205 0.24737576 0.12765465 0.0589016 0.02564742 0.00515905', 'units': 'nm', 'wavelength_max': 582.0, 'wavelength_min': 536.0}, '04': {'bandwidth': '30.0', 'central_wavelength': 665.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.3405629', 'physical_gain': '4.76622535', 'spectral_response_step': '1', 'spectral_response_values': '0.00499358 0.02642563 0.11905127 0.333204 0.59813448 0.80612041 0.91152955 0.92179127 0.91677167 0.90751672 0.89867522 0.89413622 0.89685141 0.89933396 0.90191681 0.90710817 0.9164622 0.92908702 0.9426682 0.95591935 0.96854537 0.98264967 0.99231022 1 0.99904114 0.99257339 0.97943242 0.96553214 0.95377013 0.94146127 0.92151286 0.89308475 0.83539461 0.69759082 0.49483622 0.27886075 0.10892715 0.03028701 0.00747382 0.00087148', 'units': 'nm', 'wavelength_max': 685.0, 'wavelength_min': 646.0}, '05': {'bandwidth': '15.0', 'central_wavelength': 703.8, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7846026', 'physical_gain': '5.17334731', 'spectral_response_step': '1', 'spectral_response_values': '0.01042619 0.05713826 0.21461286 0.54715702 0.87088164 0.96808183 0.99104427 1 0.99512875 0.98751456 0.97910038 0.97035979 0.95875454 0.94130694 0.92531149 0.89283152 0.76531084 0.50228771 0.17957688 0.0337948 0.00240526', 'units': 'nm', 'wavelength_max': 714.0, 'wavelength_min': 694.0}, '06': {'bandwidth': '13.0', 'central_wavelength': 739.1, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7727115', 'physical_gain': '5.08418111', 'spectral_response_step': '1', 'spectral_response_values': '0.01739744 0.10565746 0.38571484 0.78168196 0.90518378 0.91562509 0.92258804 0.93134141 0.9469604 0.96535098 0.97817455 0.99107716 0.99990615 1 0.97144118 0.81937503 0.46748011 0.09409351 0.00983236', 'units': 'nm', 'wavelength_max': 748.0, 'wavelength_min': 730.0}, '07': {'bandwidth': '19.0', 'central_wavelength': 779.7, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7727115', 'physical_gain': '4.75944225', 'spectral_response_step': '1', 'spectral_response_values': '0.0103729 0.03754921 0.11240409 0.25890105 0.48035521 0.73155954 0.91293607 0.97124929 0.96391197 0.95529249 0.964831 0.98628988 1 0.99782157 0.98343012 0.96489467 0.94619093 0.92560158 0.90788186 0.88471259 0.85693094 0.82513165 0.7734046 0.66767522 0.47756609 0.23225321 0.06764032 0.01301456 0.00117425', 'units': 'nm', 'wavelength_max': 794.0, 'wavelength_min': 766.0}, '08': {'bandwidth': '105.0', 'central_wavelength': 833.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.3872929', 'physical_gain': '6.81687988', 'spectral_response_step': '1', 'spectral_response_values': '0.00037316 0.00296451 0.01663315 0.02818619 0.05000442 0.08575595 0.14871265 0.25701156 0.42307501 0.62943997 0.80112571 0.90352196 0.93855197 0.94406104 0.9474892 0.96325767 0.97948137 0.99144397 0.99630748 1 0.99827121 0.99843182 0.98914342 0.98264167 0.96769944 0.95752283 0.95074919 0.9458125 0.94267916 0.9465958 0.94450012 0.93992861 0.92759688 0.91226544 0.89079677 0.8706102 0.85021777 0.83416655 0.82214927 0.8124078 0.80920229 0.80220847 0.79081499 0.78239761 0.76731527 0.75394962 0.74226922 0.72750987 0.71976209 0.71456726 0.71982866 0.72746214 0.73945306 0.75138424 0.76310661 0.77122498 0.78298312 0.78494127 0.78409222 0.7834498 0.78216032 0.78062401 0.78132572 0.7813272 0.7810081 0.77897938 0.7761445 0.76910534 0.7625494 0.75157186 0.74086146 0.73121299 0.71988688 0.71025573 0.69679744 0.68602501 0.67163906 0.65532408 0.64173681 0.62683353 0.61241074 0.60185411 0.59380689 0.58714687 0.58444579 0.58231388 0.58111599 0.57996902 0.57480451 0.57684802 0.57273034 0.57144461 0.56985127 0.57167225 0.57154913 0.57292235 0.57617796 0.5784908 0.58023702 0.57982619 0.57868642 0.57587451 0.56976789 0.56173136 0.55644176 0.54881732 0.54508423 0.54153848 0.54069902 0.53850959 0.53655263 0.530404 0.52068821 0.50399678 0.486513 0.46813829 0.45468861 0.44447936 0.44177056 0.44425396 0.44633078 0.43914074 0.41748156 0.3690277 0.30165803 0.23504284 0.17434599 0.12247894 0.08354059 0.05624109 0.03804368 0.02427229 0.01490577 0.00615862', 'units': 'nm', 'wavelength_max': 907.0, 'wavelength_min': 774.0}, '09': {'bandwidth': '20.0', 'central_wavelength': 943.2, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '8.031501', 'physical_gain': '9.36842662', 'spectral_response_step': '1', 'spectral_response_values': '0.0121336 0.04608767 0.15156613 0.35888361 0.60704101 0.83836043 0.93474094 0.94270146 0.95838078 0.99064712 0.99789825 1 0.98593726 0.97333604 0.95776631 0.972226 0.94856942 0.94367414 0.90771555 0.88460732 0.85258329 0.83375172 0.71599386 0.52202762 0.26922852 0.09477806 0.02640828 0.00346547', 'units': 'nm', 'wavelength_max': 957.0, 'wavelength_min': 930.0}, '10': {'bandwidth': '30.0', 'central_wavelength': 1376.9, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '5.5537686', 'physical_gain': '56.4937506', 'spectral_response_step': '1', 'spectral_response_values': '2.472e-05 0.00013691 0.00012558 8.901e-05 0.00012425 9.941e-05 0.00013952 0.00015816 0.00019272 0.00025959 0.00032221 0.00034719 0.0003699 0.00054874 0.00105434 0.00218813 0.00480743 0.01135252 0.02671185 0.05776022 0.11176337 0.19587518 0.31418191 0.46188068 0.62292578 0.7709851 0.88086652 0.9448941 0.97405066 0.98616696 0.99306955 0.99775441 1 0.99942348 0.99616891 0.99082045 0.9842131 0.97708513 0.97013647 0.96374366 0.95755001 0.95127438 0.94546638 0.94069659 0.93759595 0.93624612 0.93510206 0.93054472 0.91630845 0.88530334 0.83129653 0.74856466 0.63524397 0.49733159 0.34907723 0.21259735 0.10971453 0.04789269 0.01853013 0.00716776 0.0031533 0.00157017 0.00084901 0.00053006 0.00033171 0.00019447 0.00022104 0.00022646 0.00018156 0.00016063 0.00015475 0.00014734 0.00014776 0.00017405 0.00023619 0.00012007 4.337e-05', 'units': 'nm', 'wavelength_max': 1415.0, 'wavelength_min': 1339.0}, '11': {'bandwidth': '90.0', 'central_wavelength': 1610.4, 'onboard_compression_rate': '2.4', 'onboard_integration_time': '1.3793689', 'physical_gain': '37.19331115', 'spectral_response_step': '1', 'spectral_response_values': '1.154e-05 2.707e-05 8.129e-05 0.0001701 0.00027422 0.00034456 0.00046028 0.00065214 0.00082283 0.00107791 0.0014306 0.00196134 0.00266427 0.00368682 0.00522456 0.00758401 0.01126335 0.01715812 0.02674581 0.04145595 0.06300627 0.09464207 0.13995799 0.20105412 0.28189591 0.38134665 0.4907345 0.60674263 0.71505301 0.80391496 0.87015099 0.91645643 0.94668952 0.96391534 0.97305962 0.97704089 0.97777566 0.97686717 0.97531356 0.97336816 0.9714563 0.9697157 0.96907419 0.96968255 0.97051178 0.97272986 0.97613656 0.97894419 0.9810083 0.98350836 0.9848292 0.98438948 0.98389859 0.98334634 0.9814301 0.97936035 0.97802641 0.97623515 0.97537114 0.97569131 0.97679261 0.97898052 0.98199689 0.98520852 0.98866135 0.99233425 0.99480248 0.99589079 0.9958911 0.99475534 0.99207775 0.98856394 0.9848769 0.98106836 0.97677436 0.97351815 0.97192459 0.97052192 0.97043004 0.9723835 0.97525347 0.97856769 0.98298866 0.98810437 0.99268138 0.99645012 0.9990686 1 0.99902738 0.99662493 0.99326995 0.989647 0.98577051 0.98212932 0.97979728 0.97946062 0.980262 0.98247241 0.98601349 0.98957829 0.99173488 0.99219848 0.98937107 0.98205611 0.97007817 0.95307506 0.93223131 0.90784439 0.88392149 0.86411672 0.85075738 0.84410342 0.84337963 0.84698191 0.84866039 0.84046041 0.81336359 0.75654857 0.66994259 0.56517119 0.45690882 0.35310835 0.25633426 0.17582806 0.11552613 0.0733101 0.04640345 0.02898639 0.01853597 0.01243537 0.00877131 0.00630418 0.00457459 0.00335323 0.00245906 0.001988 0.00149989 0.00112208 0.00078208 0.00054086 0.00028019 0.0001326', 'units': 'nm', 'wavelength_max': 1679.0, 'wavelength_min': 1538.0}, '12': {'bandwidth': '180.0', 'central_wavelength': 2185.7, 'onboard_compression_rate': '2.4', 'onboard_integration_time': '1.4761667', 'physical_gain': '108.59180072', 'spectral_response_step': '1', 'spectral_response_values': '0.00022389 0.00073676 0.00164703 0.00301151 0.00458328 0.00592584 0.00752876 0.00874103 0.01025764 0.01222618 0.01458055 0.01744267 0.02104287 0.02540339 0.03057901 0.03719619 0.04572365 0.05630242 0.06994211 0.08791078 0.11057655 0.13873936 0.17311239 0.21416774 0.26175285 0.31696031 0.38057337 0.44916129 0.52246923 0.59858476 0.67183039 0.73762307 0.79267856 0.83543144 0.86612544 0.88613168 0.89739036 0.90131058 0.90056883 0.89688046 0.89211284 0.88771935 0.88417799 0.88183136 0.88142338 0.88242075 0.88687585 0.89245189 0.89861914 0.90533051 0.91254666 0.91988456 0.92662076 0.93280462 0.9381479 0.94308713 0.94528987 0.94711578 0.94827846 0.94854335 0.94782599 0.94683272 0.94584552 0.94458923 0.94320642 0.94161778 0.94216937 0.94300085 0.94396863 0.94500784 0.94593652 0.94680905 0.94766521 0.94866638 0.94960932 0.95040536 0.95078607 0.95126357 0.95205865 0.9524548 0.95228734 0.95215614 0.95239704 0.95270563 0.95306455 0.9535262 0.95404061 0.9545903 0.95529443 0.95650666 0.95774374 0.95899449 0.96021128 0.96117558 0.96241242 0.96389292 0.9633817 0.96287807 0.96252982 0.96163134 0.96051578 0.95934879 0.95816596 0.95676127 0.95491383 0.95287555 0.95386662 0.95498503 0.9555239 0.95548083 0.9551674 0.95442758 0.95327341 0.95194429 0.95041866 0.9484181 0.94839027 0.94838056 0.94813039 0.94753901 0.94645196 0.94504703 0.9433144 0.94197531 0.94049427 0.93875727 0.94132922 0.9436129 0.94558364 0.94711284 0.94831802 0.94945878 0.95060227 0.95182631 0.95345901 0.95510105 0.95225316 0.95033203 0.94956701 0.94895037 0.94955832 0.95152282 0.95486528 0.95963437 0.96607045 0.97375116 0.97078626 0.96900558 0.96819786 0.96750837 0.9674355 0.96792378 0.96839035 0.96883692 0.9693902 0.96985301 0.96898817 0.96814874 0.96743874 0.96638941 0.96534457 0.96425351 0.96281969 0.96155971 0.96053251 0.95926107 0.95959913 0.96025143 0.96154033 0.96262988 0.96359875 0.96467154 0.9654114 0.96625109 0.96744643 0.96878244 0.97046916 0.97234778 0.97422228 0.9759326 0.97713045 0.97892333 0.98039008 0.98147316 0.98248415 0.9832885 0.98458694 0.9866985 0.98911057 0.99119702 0.99315819 0.99524701 0.99668121 0.99826512 0.99959594 1 0.99866374 0.99576531 0.99062502 0.98155399 0.96876193 0.95174168 0.92942389 0.90240499 0.8705554 0.83207693 0.78626172 0.73649625 0.68202115 0.62329799 0.56418429 0.50680538 0.4515209 0.4006232 0.35430248 0.30995212 0.26941268 0.23418861 0.20232924 0.17373524 0.14941215 0.1290062 0.11157271 0.09693492 0.08490613 0.07409041 0.06449244 0.056343 0.0493999 0.04321207 0.03800099 0.03348242 0.02900748 0.0251013 0.02003645 0.01384563 0.00850471 0.00443484 0.00085324', 'units': 'nm', 'wavelength_max': 2303.0, 'wavelength_min': 2065.0}, '8A': {'bandwidth': '20.0', 'central_wavelength': 864.0, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7608204', 'physical_gain': '5.7570522', 'spectral_response_step': '1', 'spectral_response_values': '0.00167523 0.01602231 0.03225867 0.07345268 0.1689243 0.34543042 0.56923369 0.79611745 0.93749188 0.98102805 0.98742384 0.99457226 0.99912415 0.99993652 1 0.99437257 0.98756135 0.98263615 0.9790323 0.97397518 0.97130259 0.9645338 0.95610202 0.93941552 0.89155652 0.77601041 0.5951886 0.37588812 0.18394037 0.07870072 0.0332686 0.01575167 0.00159818', 'units': 'nm', 'wavelength_max': 880.0, 'wavelength_min': 848.0}, 'b01': {'bandwidth': 20.0, 'central_wavelength': 442.3, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '7.22833', 'physical_gain': '3.95548343', 'spectral_response_step': '1', 'spectral_response_values': '0.0062411 0.01024045 0.00402983 0.00642179 0.00552753 0.0065525 0.00409887 0.006297 0.00436742 0.00233356 0.00058162 0.00202276 0.00294328 0.00485362 0.00317041 0.00237657 0.00234612 0.00440152 0.01292397 0.05001678 0.18650104 0.45441623 0.72307877 0.83999211 0.86456334 0.87472096 0.89215296 0.91090814 0.92588017 0.93924094 0.94491826 0.95078529 0.96803023 0.99939195 1 0.97548364 0.96148351 0.94986211 0.91841452 0.87989802 0.80383677 0.59752075 0.30474132 0.10798014 0.0304465 0.00885119', 'units': 'nm', 'wavelength_max': 456.0, 'wavelength_min': 411.0}, 'b02': {'bandwidth': 65.0, 'central_wavelength': 492.3, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.2646269', 'physical_gain': '3.81250879', 'spectral_response_step': '1', 'spectral_response_values': '0.05529541 0.12005068 0.25199051 0.4623617 0.65162379 0.77642171 0.82319091 0.83083116 0.83382106 0.837526 0.86304286 0.88226141 0.90486326 0.92043837 0.93602675 0.930533 0.92714067 0.9161479 0.90551724 0.89745515 0.90266694 0.90854264 0.92047913 0.92417935 0.91845025 0.90743244 0.89733983 0.88646415 0.87189983 0.85643973 0.84473414 0.84190734 0.85644111 0.87782724 0.90261174 0.91840544 0.94585847 0.96887192 0.99336135 0.99927899 1 0.99520325 0.98412711 0.97947473 0.97808297 0.97213439 0.96277794 0.95342234 0.93802376 0.92460144 0.90932642 0.90192251 0.89184298 0.88963556 0.89146958 0.89877911 0.91056869 0.92427362 0.93823555 0.95311791 0.97150808 0.98737003 0.99658514 0.99367959 0.98144714 0.95874415 0.89291635 0.73566218 0.52060373 0.3322804 0.19492197 0.11732617 0.07507304 0.05094154 0.03213016 0.01510217 0.00447984', 'units': 'nm', 'wavelength_max': 532.0, 'wavelength_min': 456.0}, 'b03': {'bandwidth': 35.0, 'central_wavelength': 559.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.2996743', 'physical_gain': '4.22866877', 'spectral_response_step': '1', 'spectral_response_values': '0.00188039 0.01545903 0.03660414 0.08100583 0.16917887 0.33278274 0.58622794 0.80916412 0.913051 0.94472284 0.94898813 0.94369132 0.92845674 0.91256938 0.90078036 0.89958598 0.90547138 0.92045355 0.94065665 0.96199681 0.98186744 0.9985841 1 0.99279888 0.97801325 0.95301174 0.9266333 0.89359131 0.86941793 0.84827 0.83908301 0.83206209 0.8291787 0.83305844 0.84630939 0.86396307 0.87268076 0.86818339 0.8554947 0.80839054 0.67650876 0.45584205 0.24737576 0.12765465 0.0589016 0.02564742 0.00515905', 'units': 'nm', 'wavelength_max': 582.0, 'wavelength_min': 536.0}, 'b04': {'bandwidth': 30.0, 'central_wavelength': 665.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.3405629', 'physical_gain': '4.76622535', 'spectral_response_step': '1', 'spectral_response_values': '0.00499358 0.02642563 0.11905127 0.333204 0.59813448 0.80612041 0.91152955 0.92179127 0.91677167 0.90751672 0.89867522 0.89413622 0.89685141 0.89933396 0.90191681 0.90710817 0.9164622 0.92908702 0.9426682 0.95591935 0.96854537 0.98264967 0.99231022 1 0.99904114 0.99257339 0.97943242 0.96553214 0.95377013 0.94146127 0.92151286 0.89308475 0.83539461 0.69759082 0.49483622 0.27886075 0.10892715 0.03028701 0.00747382 0.00087148', 'units': 'nm', 'wavelength_max': 685.0, 'wavelength_min': 646.0}, 'b05': {'bandwidth': 15.0, 'central_wavelength': 703.8, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7846026', 'physical_gain': '5.17334731', 'spectral_response_step': '1', 'spectral_response_values': '0.01042619 0.05713826 0.21461286 0.54715702 0.87088164 0.96808183 0.99104427 1 0.99512875 0.98751456 0.97910038 0.97035979 0.95875454 0.94130694 0.92531149 0.89283152 0.76531084 0.50228771 0.17957688 0.0337948 0.00240526', 'units': 'nm', 'wavelength_max': 714.0, 'wavelength_min': 694.0}, 'b06': {'bandwidth': 13.0, 'central_wavelength': 739.1, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7727115', 'physical_gain': '5.08418111', 'spectral_response_step': '1', 'spectral_response_values': '0.01739744 0.10565746 0.38571484 0.78168196 0.90518378 0.91562509 0.92258804 0.93134141 0.9469604 0.96535098 0.97817455 0.99107716 0.99990615 1 0.97144118 0.81937503 0.46748011 0.09409351 0.00983236', 'units': 'nm', 'wavelength_max': 748.0, 'wavelength_min': 730.0}, 'b07': {'bandwidth': 19.0, 'central_wavelength': 779.7, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7727115', 'physical_gain': '4.75944225', 'spectral_response_step': '1', 'spectral_response_values': '0.0103729 0.03754921 0.11240409 0.25890105 0.48035521 0.73155954 0.91293607 0.97124929 0.96391197 0.95529249 0.964831 0.98628988 1 0.99782157 0.98343012 0.96489467 0.94619093 0.92560158 0.90788186 0.88471259 0.85693094 0.82513165 0.7734046 0.66767522 0.47756609 0.23225321 0.06764032 0.01301456 0.00117425', 'units': 'nm', 'wavelength_max': 794.0, 'wavelength_min': 766.0}, 'b08': {'bandwidth': 105.0, 'central_wavelength': 833.0, 'onboard_compression_rate': '2.97', 'onboard_integration_time': '1.3872929', 'physical_gain': '6.81687988', 'spectral_response_step': '1', 'spectral_response_values': '0.00037316 0.00296451 0.01663315 0.02818619 0.05000442 0.08575595 0.14871265 0.25701156 0.42307501 0.62943997 0.80112571 0.90352196 0.93855197 0.94406104 0.9474892 0.96325767 0.97948137 0.99144397 0.99630748 1 0.99827121 0.99843182 0.98914342 0.98264167 0.96769944 0.95752283 0.95074919 0.9458125 0.94267916 0.9465958 0.94450012 0.93992861 0.92759688 0.91226544 0.89079677 0.8706102 0.85021777 0.83416655 0.82214927 0.8124078 0.80920229 0.80220847 0.79081499 0.78239761 0.76731527 0.75394962 0.74226922 0.72750987 0.71976209 0.71456726 0.71982866 0.72746214 0.73945306 0.75138424 0.76310661 0.77122498 0.78298312 0.78494127 0.78409222 0.7834498 0.78216032 0.78062401 0.78132572 0.7813272 0.7810081 0.77897938 0.7761445 0.76910534 0.7625494 0.75157186 0.74086146 0.73121299 0.71988688 0.71025573 0.69679744 0.68602501 0.67163906 0.65532408 0.64173681 0.62683353 0.61241074 0.60185411 0.59380689 0.58714687 0.58444579 0.58231388 0.58111599 0.57996902 0.57480451 0.57684802 0.57273034 0.57144461 0.56985127 0.57167225 0.57154913 0.57292235 0.57617796 0.5784908 0.58023702 0.57982619 0.57868642 0.57587451 0.56976789 0.56173136 0.55644176 0.54881732 0.54508423 0.54153848 0.54069902 0.53850959 0.53655263 0.530404 0.52068821 0.50399678 0.486513 0.46813829 0.45468861 0.44447936 0.44177056 0.44425396 0.44633078 0.43914074 0.41748156 0.3690277 0.30165803 0.23504284 0.17434599 0.12247894 0.08354059 0.05624109 0.03804368 0.02427229 0.01490577 0.00615862', 'units': 'nm', 'wavelength_max': 907.0, 'wavelength_min': 774.0}, 'b09': {'bandwidth': 20.0, 'central_wavelength': 943.2, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '8.031501', 'physical_gain': '9.36842662', 'spectral_response_step': '1', 'spectral_response_values': '0.0121336 0.04608767 0.15156613 0.35888361 0.60704101 0.83836043 0.93474094 0.94270146 0.95838078 0.99064712 0.99789825 1 0.98593726 0.97333604 0.95776631 0.972226 0.94856942 0.94367414 0.90771555 0.88460732 0.85258329 0.83375172 0.71599386 0.52202762 0.26922852 0.09477806 0.02640828 0.00346547', 'units': 'nm', 'wavelength_max': 957.0, 'wavelength_min': 930.0}, 'b10': {'bandwidth': 30.0, 'central_wavelength': 1376.9, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '5.5537686', 'physical_gain': '56.4937506', 'spectral_response_step': '1', 'spectral_response_values': '2.472e-05 0.00013691 0.00012558 8.901e-05 0.00012425 9.941e-05 0.00013952 0.00015816 0.00019272 0.00025959 0.00032221 0.00034719 0.0003699 0.00054874 0.00105434 0.00218813 0.00480743 0.01135252 0.02671185 0.05776022 0.11176337 0.19587518 0.31418191 0.46188068 0.62292578 0.7709851 0.88086652 0.9448941 0.97405066 0.98616696 0.99306955 0.99775441 1 0.99942348 0.99616891 0.99082045 0.9842131 0.97708513 0.97013647 0.96374366 0.95755001 0.95127438 0.94546638 0.94069659 0.93759595 0.93624612 0.93510206 0.93054472 0.91630845 0.88530334 0.83129653 0.74856466 0.63524397 0.49733159 0.34907723 0.21259735 0.10971453 0.04789269 0.01853013 0.00716776 0.0031533 0.00157017 0.00084901 0.00053006 0.00033171 0.00019447 0.00022104 0.00022646 0.00018156 0.00016063 0.00015475 0.00014734 0.00014776 0.00017405 0.00023619 0.00012007 4.337e-05', 'units': 'nm', 'wavelength_max': 1415.0, 'wavelength_min': 1339.0}, 'b11': {'bandwidth': 90.0, 'central_wavelength': 1610.4, 'onboard_compression_rate': '2.4', 'onboard_integration_time': '1.3793689', 'physical_gain': '37.19331115', 'spectral_response_step': '1', 'spectral_response_values': '1.154e-05 2.707e-05 8.129e-05 0.0001701 0.00027422 0.00034456 0.00046028 0.00065214 0.00082283 0.00107791 0.0014306 0.00196134 0.00266427 0.00368682 0.00522456 0.00758401 0.01126335 0.01715812 0.02674581 0.04145595 0.06300627 0.09464207 0.13995799 0.20105412 0.28189591 0.38134665 0.4907345 0.60674263 0.71505301 0.80391496 0.87015099 0.91645643 0.94668952 0.96391534 0.97305962 0.97704089 0.97777566 0.97686717 0.97531356 0.97336816 0.9714563 0.9697157 0.96907419 0.96968255 0.97051178 0.97272986 0.97613656 0.97894419 0.9810083 0.98350836 0.9848292 0.98438948 0.98389859 0.98334634 0.9814301 0.97936035 0.97802641 0.97623515 0.97537114 0.97569131 0.97679261 0.97898052 0.98199689 0.98520852 0.98866135 0.99233425 0.99480248 0.99589079 0.9958911 0.99475534 0.99207775 0.98856394 0.9848769 0.98106836 0.97677436 0.97351815 0.97192459 0.97052192 0.97043004 0.9723835 0.97525347 0.97856769 0.98298866 0.98810437 0.99268138 0.99645012 0.9990686 1 0.99902738 0.99662493 0.99326995 0.989647 0.98577051 0.98212932 0.97979728 0.97946062 0.980262 0.98247241 0.98601349 0.98957829 0.99173488 0.99219848 0.98937107 0.98205611 0.97007817 0.95307506 0.93223131 0.90784439 0.88392149 0.86411672 0.85075738 0.84410342 0.84337963 0.84698191 0.84866039 0.84046041 0.81336359 0.75654857 0.66994259 0.56517119 0.45690882 0.35310835 0.25633426 0.17582806 0.11552613 0.0733101 0.04640345 0.02898639 0.01853597 0.01243537 0.00877131 0.00630418 0.00457459 0.00335323 0.00245906 0.001988 0.00149989 0.00112208 0.00078208 0.00054086 0.00028019 0.0001326', 'units': 'nm', 'wavelength_max': 1679.0, 'wavelength_min': 1538.0}, 'b12': {'bandwidth': 180.0, 'central_wavelength': 2185.7, 'onboard_compression_rate': '2.4', 'onboard_integration_time': '1.4761667', 'physical_gain': '108.59180072', 'spectral_response_step': '1', 'spectral_response_values': '0.00022389 0.00073676 0.00164703 0.00301151 0.00458328 0.00592584 0.00752876 0.00874103 0.01025764 0.01222618 0.01458055 0.01744267 0.02104287 0.02540339 0.03057901 0.03719619 0.04572365 0.05630242 0.06994211 0.08791078 0.11057655 0.13873936 0.17311239 0.21416774 0.26175285 0.31696031 0.38057337 0.44916129 0.52246923 0.59858476 0.67183039 0.73762307 0.79267856 0.83543144 0.86612544 0.88613168 0.89739036 0.90131058 0.90056883 0.89688046 0.89211284 0.88771935 0.88417799 0.88183136 0.88142338 0.88242075 0.88687585 0.89245189 0.89861914 0.90533051 0.91254666 0.91988456 0.92662076 0.93280462 0.9381479 0.94308713 0.94528987 0.94711578 0.94827846 0.94854335 0.94782599 0.94683272 0.94584552 0.94458923 0.94320642 0.94161778 0.94216937 0.94300085 0.94396863 0.94500784 0.94593652 0.94680905 0.94766521 0.94866638 0.94960932 0.95040536 0.95078607 0.95126357 0.95205865 0.9524548 0.95228734 0.95215614 0.95239704 0.95270563 0.95306455 0.9535262 0.95404061 0.9545903 0.95529443 0.95650666 0.95774374 0.95899449 0.96021128 0.96117558 0.96241242 0.96389292 0.9633817 0.96287807 0.96252982 0.96163134 0.96051578 0.95934879 0.95816596 0.95676127 0.95491383 0.95287555 0.95386662 0.95498503 0.9555239 0.95548083 0.9551674 0.95442758 0.95327341 0.95194429 0.95041866 0.9484181 0.94839027 0.94838056 0.94813039 0.94753901 0.94645196 0.94504703 0.9433144 0.94197531 0.94049427 0.93875727 0.94132922 0.9436129 0.94558364 0.94711284 0.94831802 0.94945878 0.95060227 0.95182631 0.95345901 0.95510105 0.95225316 0.95033203 0.94956701 0.94895037 0.94955832 0.95152282 0.95486528 0.95963437 0.96607045 0.97375116 0.97078626 0.96900558 0.96819786 0.96750837 0.9674355 0.96792378 0.96839035 0.96883692 0.9693902 0.96985301 0.96898817 0.96814874 0.96743874 0.96638941 0.96534457 0.96425351 0.96281969 0.96155971 0.96053251 0.95926107 0.95959913 0.96025143 0.96154033 0.96262988 0.96359875 0.96467154 0.9654114 0.96625109 0.96744643 0.96878244 0.97046916 0.97234778 0.97422228 0.9759326 0.97713045 0.97892333 0.98039008 0.98147316 0.98248415 0.9832885 0.98458694 0.9866985 0.98911057 0.99119702 0.99315819 0.99524701 0.99668121 0.99826512 0.99959594 1 0.99866374 0.99576531 0.99062502 0.98155399 0.96876193 0.95174168 0.92942389 0.90240499 0.8705554 0.83207693 0.78626172 0.73649625 0.68202115 0.62329799 0.56418429 0.50680538 0.4515209 0.4006232 0.35430248 0.30995212 0.26941268 0.23418861 0.20232924 0.17373524 0.14941215 0.1290062 0.11157271 0.09693492 0.08490613 0.07409041 0.06449244 0.056343 0.0493999 0.04321207 0.03800099 0.03348242 0.02900748 0.0251013 0.02003645 0.01384563 0.00850471 0.00443484 0.00085324', 'units': 'nm', 'wavelength_max': 2303.0, 'wavelength_min': 2065.0}, 'b8a': {'bandwidth': 20.0, 'central_wavelength': 864.0, 'onboard_compression_rate': '2.655', 'onboard_integration_time': '2.7608204', 'physical_gain': '5.7570522', 'spectral_response_step': '1', 'spectral_response_values': '0.00167523 0.01602231 0.03225867 0.07345268 0.1689243 0.34543042 0.56923369 0.79611745 0.93749188 0.98102805 0.98742384 0.99457226 0.99912415 0.99993652 1 0.99437257 0.98756135 0.98263615 0.9790323 0.97397518 0.97130259 0.9645338 0.95610202 0.93941552 0.89155652 0.77601041 0.5951886 0.37588812 0.18394037 0.07870072 0.0332686 0.01575167 0.00159818', 'units': 'nm', 'wavelength_max': 880.0, 'wavelength_min': 848.0}}, 'declared_accuracy_of_AOT_model': 0.0, 'declared_accuracy_of_radiative_transfer_model': 0.0, 'declared_accuracy_of_water_vapour_model': 0.0, 'electronic_crosstalk_correction_flag': 'null', 'eopf_category': 'eoproduct', 'geometric_refinement': {'mean_value_of_residual_displacements_at_all_tie_points_after_refinement_m': {'x_mean': 'null', 'y_mean': 'null'}, 'spacecraft_rotation': {'X': {'coefficients': 'null', 'degree': 'null'}, 'Y': {'coefficients': 'null', 'degree': 'null'}, 'Z': {'coefficients': 'null', 'degree': 'null'}}, 'standard_deviation_of_residual_displacements_at_all_tie_points_after_refinement_m': {'x_stdv': 'null', 'y_stdv': 'null'}}, 'history': [{'output': 'Downlinked Stream', 'type': 'Raw Data'}, {'inputs': 'Downlinked Stream', 'organisation': 'ESA', 'output': 'S2MSIL0__etc', 'processor': 'L0', 'type': 'Level-0 Product'}, {'inputs': {'Level-0 Product': 'S2MSIL0__etc', 'list of used processing parameters file names': 'S2B_OPER_GIP_PROBAS_MPC__20240717T000511_V20240723T070000_21000101T000000_B00,S2B_OPER_GIP_ATMIMA_MPC__20170206T103051_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_INVLOC_MPC__20170523T080300_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_LREXTR_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_OLQCPA_MPC__20250210T000044_V20250211T000000_21000101T000000_B00,S2B_OPER_GIP_ATMSAD_MPC__20170324T155501_V20170306T000000_21000101T000000_B00,S2B_OPER_GIP_BLINDP_MPC__20170221T000000_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_CLOINV_MPC__20210609T000002_V20210823T030000_21000101T000000_B00,S2B_OPER_GIP_CLOPAR_MPC__20220120T000001_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_CONVER_MPC__20150710T131444_V20150627T000000_21000101T000000_B00,S2B_OPER_GIP_DATATI_MPC__20170428T123038_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_DECOMP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2__OPER_GIP_EARMOD_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_ECMWFP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2B_OPER_GIP_G2PARA_MPC__20250128T000031_V20250130T001500_21000101T000000_B00,S2B_OPER_GIP_G2PARE_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_GEOPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_HRTPAR_MPC__20221206T000000_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_INTDET_MPC__20220120T000010_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_JP2KPA_MPC__20220120T000006_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_MASPAR_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_PRDLOC_MPC__20180301T130000_V20180305T014000_21000101T000000_B00,S2B_OPER_GIP_R2ABCA_MPC__20250416T140000_V20250417T000000_21000101T000000_B00,S2B_OPER_GIP_R2BINN_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_R2CRCO_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEPI_MPC__20250312T000008_V20250313T000000_21000101T000000_B00,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B12,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B03,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B05,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B08,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B04,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B10,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B01,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B06,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B09,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B02,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B8A,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B07,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B09,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B10,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B08,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B02,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B01,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B03,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B05,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B8A,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B04,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B12,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B06,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2NOMO_MPC__20250117T000002_V20250121T080000_21000101T000000_B00,S2B_OPER_GIP_R2PARA_MPC__20221206T000009_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_R2SWIR_MPC__20170523T080300_V20170517T090600_21000101T000000_B00,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_RESPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_SPAMOD_MPC__20250428T000037_V20250430T000000_21000101T000000_B00,S2B_OPER_GIP_TILPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B8A,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B01,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B11,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B08,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B03,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B07,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B02,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B12,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B04,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B06,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B09,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B05,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B10,S2__OPER_GIP_L2ACSC_MPC__20220121T000003_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_L2ACAC_MPC__20220121T000004_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_PROBA2_MPC__20240809T000511_V20240813T070000_21000101T000000_B00', 'used CAMS file names': 'S2__OPER_AUX_CAMSFO_ADG__20250430T000000_V20250430T000000_20250502T010000', 'used DEM file name': 'CopernicusDEM30', 'used ECMWF file names': 'S2__OPER_AUX_ECMWFD_ADG__20250430T000000_V20250430T090000_20250502T030000', 'used GRI file name': 'S2A_OPER_GRI_MSIL1B_MPC__20160521T184422_S20151114T103133,S2A_OPER_GRI_MSIL1B_MPC__20160531T210152_S20160512T102954,S2A_OPER_GRI_MSIL1B_MPC__20160907T095549_S20160422T103554,S2A_OPER_GRI_MSIL1B_MPC__20161018T120000_S20150806T102429,S2A_OPER_GRI_MSIL1B_MPC__20161018T120000_S20150826T102633', 'used IERS file name': 'S2__OPER_AUX_UT1UTC_PDMC_20250424T000000_V20250425T000000_20260424T000000'}, 'organisation': 'ESA', 'output': 'S2MSIL1A_etc', 'processor': 'Sentinel-2 IPF', 'type': 'Level-1A Product', 'version': '???'}, {'inputs': {'Level-1A Product': 'S2MSIL1A_etc', 'list of used processing parameters file names': 'S2B_OPER_GIP_PROBAS_MPC__20240717T000511_V20240723T070000_21000101T000000_B00,S2B_OPER_GIP_ATMIMA_MPC__20170206T103051_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_INVLOC_MPC__20170523T080300_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_LREXTR_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_OLQCPA_MPC__20250210T000044_V20250211T000000_21000101T000000_B00,S2B_OPER_GIP_ATMSAD_MPC__20170324T155501_V20170306T000000_21000101T000000_B00,S2B_OPER_GIP_BLINDP_MPC__20170221T000000_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_CLOINV_MPC__20210609T000002_V20210823T030000_21000101T000000_B00,S2B_OPER_GIP_CLOPAR_MPC__20220120T000001_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_CONVER_MPC__20150710T131444_V20150627T000000_21000101T000000_B00,S2B_OPER_GIP_DATATI_MPC__20170428T123038_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_DECOMP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2__OPER_GIP_EARMOD_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_ECMWFP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2B_OPER_GIP_G2PARA_MPC__20250128T000031_V20250130T001500_21000101T000000_B00,S2B_OPER_GIP_G2PARE_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_GEOPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_HRTPAR_MPC__20221206T000000_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_INTDET_MPC__20220120T000010_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_JP2KPA_MPC__20220120T000006_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_MASPAR_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_PRDLOC_MPC__20180301T130000_V20180305T014000_21000101T000000_B00,S2B_OPER_GIP_R2ABCA_MPC__20250416T140000_V20250417T000000_21000101T000000_B00,S2B_OPER_GIP_R2BINN_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_R2CRCO_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEPI_MPC__20250312T000008_V20250313T000000_21000101T000000_B00,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B12,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B03,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B05,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B08,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B04,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B10,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B01,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B06,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B09,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B02,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B8A,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B07,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B09,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B10,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B08,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B02,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B01,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B03,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B05,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B8A,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B04,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B12,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B06,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2NOMO_MPC__20250117T000002_V20250121T080000_21000101T000000_B00,S2B_OPER_GIP_R2PARA_MPC__20221206T000009_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_R2SWIR_MPC__20170523T080300_V20170517T090600_21000101T000000_B00,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_RESPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_SPAMOD_MPC__20250428T000037_V20250430T000000_21000101T000000_B00,S2B_OPER_GIP_TILPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B8A,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B01,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B11,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B08,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B03,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B07,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B02,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B12,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B04,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B06,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B09,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B05,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B10,S2__OPER_GIP_L2ACSC_MPC__20220121T000003_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_L2ACAC_MPC__20220121T000004_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_PROBA2_MPC__20240809T000511_V20240813T070000_21000101T000000_B00', 'used CAMS file names': 'S2__OPER_AUX_CAMSFO_ADG__20250430T000000_V20250430T000000_20250502T010000', 'used DEM file name': 'CopernicusDEM30', 'used ECMWF file names': 'S2__OPER_AUX_ECMWFD_ADG__20250430T000000_V20250430T090000_20250502T030000', 'used GRI file name': 'S2A_OPER_GRI_MSIL1B_MPC__20160521T184422_S20151114T103133,S2A_OPER_GRI_MSIL1B_MPC__20160531T210152_S20160512T102954,S2A_OPER_GRI_MSIL1B_MPC__20160907T095549_S20160422T103554,S2A_OPER_GRI_MSIL1B_MPC__20161018T120000_S20150806T102429,S2A_OPER_GRI_MSIL1B_MPC__20161018T120000_S20150826T102633', 'used IERS file name': 'S2__OPER_AUX_UT1UTC_PDMC_20250424T000000_V20250425T000000_20260424T000000'}, 'organisation': 'ESA', 'output': 'S2MSIL1B_etc', 'processor': 'Sentinel-2 IPF', 'type': 'Level-1B Product', 'version': '???'}, {'inputs': {'Level-1B Product': 'S2MSIL1B_etc', 'list of used processing parameters file names': 'S2B_OPER_GIP_PROBAS_MPC__20240717T000511_V20240723T070000_21000101T000000_B00,S2B_OPER_GIP_ATMIMA_MPC__20170206T103051_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_INVLOC_MPC__20170523T080300_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_LREXTR_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_OLQCPA_MPC__20250210T000044_V20250211T000000_21000101T000000_B00,S2B_OPER_GIP_ATMSAD_MPC__20170324T155501_V20170306T000000_21000101T000000_B00,S2B_OPER_GIP_BLINDP_MPC__20170221T000000_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_CLOINV_MPC__20210609T000002_V20210823T030000_21000101T000000_B00,S2B_OPER_GIP_CLOPAR_MPC__20220120T000001_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_CONVER_MPC__20150710T131444_V20150627T000000_21000101T000000_B00,S2B_OPER_GIP_DATATI_MPC__20170428T123038_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_DECOMP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2__OPER_GIP_EARMOD_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_ECMWFP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2B_OPER_GIP_G2PARA_MPC__20250128T000031_V20250130T001500_21000101T000000_B00,S2B_OPER_GIP_G2PARE_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_GEOPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_HRTPAR_MPC__20221206T000000_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_INTDET_MPC__20220120T000010_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_JP2KPA_MPC__20220120T000006_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_MASPAR_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_PRDLOC_MPC__20180301T130000_V20180305T014000_21000101T000000_B00,S2B_OPER_GIP_R2ABCA_MPC__20250416T140000_V20250417T000000_21000101T000000_B00,S2B_OPER_GIP_R2BINN_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_R2CRCO_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEPI_MPC__20250312T000008_V20250313T000000_21000101T000000_B00,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B12,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B03,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B05,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B08,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B04,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B10,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B01,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B06,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B09,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B02,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B8A,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B07,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B09,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B10,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B08,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B02,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B01,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B03,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B05,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B8A,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B04,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B12,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B06,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2NOMO_MPC__20250117T000002_V20250121T080000_21000101T000000_B00,S2B_OPER_GIP_R2PARA_MPC__20221206T000009_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_R2SWIR_MPC__20170523T080300_V20170517T090600_21000101T000000_B00,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_RESPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_SPAMOD_MPC__20250428T000037_V20250430T000000_21000101T000000_B00,S2B_OPER_GIP_TILPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B8A,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B01,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B11,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B08,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B03,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B07,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B02,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B12,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B04,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B06,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B09,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B05,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B10,S2__OPER_GIP_L2ACSC_MPC__20220121T000003_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_L2ACAC_MPC__20220121T000004_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_PROBA2_MPC__20240809T000511_V20240813T070000_21000101T000000_B00', 'used CAMS file names': 'S2__OPER_AUX_CAMSFO_ADG__20250430T000000_V20250430T000000_20250502T010000', 'used DEM file name': 'CopernicusDEM30', 'used ECMWF file names': 'S2__OPER_AUX_ECMWFD_ADG__20250430T000000_V20250430T090000_20250502T030000', 'used GRI file name': 'S2A_OPER_GRI_MSIL1B_MPC__20160521T184422_S20151114T103133,S2A_OPER_GRI_MSIL1B_MPC__20160531T210152_S20160512T102954,S2A_OPER_GRI_MSIL1B_MPC__20160907T095549_S20160422T103554,S2A_OPER_GRI_MSIL1B_MPC__20161018T120000_S20150806T102429,S2A_OPER_GRI_MSIL1B_MPC__20161018T120000_S20150826T102633', 'used IERS file name': 'S2__OPER_AUX_UT1UTC_PDMC_20250424T000000_V20250425T000000_20260424T000000'}, 'organisation': 'ESA', 'output': 'S2MSIL1C_etc', 'processor': 'Sentinel-2 IPF', 'type': 'Level-1C Product', 'version': '???'}, {'inputs': {'Level-1C Product': 'S2MSIL1C_etc', 'list of used processing parameters file names': 'S2B_OPER_GIP_PROBAS_MPC__20240717T000511_V20240723T070000_21000101T000000_B00,S2B_OPER_GIP_ATMIMA_MPC__20170206T103051_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_INVLOC_MPC__20170523T080300_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_LREXTR_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_OLQCPA_MPC__20250210T000044_V20250211T000000_21000101T000000_B00,S2B_OPER_GIP_ATMSAD_MPC__20170324T155501_V20170306T000000_21000101T000000_B00,S2B_OPER_GIP_BLINDP_MPC__20170221T000000_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_CLOINV_MPC__20210609T000002_V20210823T030000_21000101T000000_B00,S2B_OPER_GIP_CLOPAR_MPC__20220120T000001_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_CONVER_MPC__20150710T131444_V20150627T000000_21000101T000000_B00,S2B_OPER_GIP_DATATI_MPC__20170428T123038_V20170322T000000_21000101T000000_B00,S2B_OPER_GIP_DECOMP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2__OPER_GIP_EARMOD_MPC__20210608T000001_V20150622T000000_21000101T000000_B00,S2B_OPER_GIP_ECMWFP_MPC__20121031T075922_V19830101T000000_21000101T000000_B00,S2B_OPER_GIP_G2PARA_MPC__20250128T000031_V20250130T001500_21000101T000000_B00,S2B_OPER_GIP_G2PARE_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_GEOPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_HRTPAR_MPC__20221206T000000_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_INTDET_MPC__20220120T000010_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_JP2KPA_MPC__20220120T000006_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_MASPAR_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_PRDLOC_MPC__20180301T130000_V20180305T014000_21000101T000000_B00,S2B_OPER_GIP_R2ABCA_MPC__20250416T140000_V20250417T000000_21000101T000000_B00,S2B_OPER_GIP_R2BINN_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_R2CRCO_MPC__20220120T000002_V20220125T022000_21000101T000000_B00,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DECT_MPC__20170206T103038_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEFI_MPC__20170206T103038_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DEFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DEFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2DENT_MPC__20170206T103039_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2DENT_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2DEPI_MPC__20250312T000008_V20250313T000000_21000101T000000_B00,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B12,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B03,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B05,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B08,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B04,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B10,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B01,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B06,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B09,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B02,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B8A,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B07,S2B_OPER_GIP_R2EOB2_MPC__20170517T113826_V20170101T000005_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B09,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B10,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B11,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B08,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B02,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B01,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B03,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B05,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B8A,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B04,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B12,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B06,S2B_OPER_GIP_R2EQOG_MPC__20250416T140000_V20250417T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2L2NC_MPC__20170206T103039_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2L2NC_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2NOMO_MPC__20250117T000002_V20250121T080000_21000101T000000_B00,S2B_OPER_GIP_R2PARA_MPC__20221206T000009_V20221206T064000_21000101T000000_B00,S2B_OPER_GIP_R2SWIR_MPC__20170523T080300_V20170517T090600_21000101T000000_B00,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B02,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B04,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B10,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B8A,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B01,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B07,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B06,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B09,S2B_OPER_GIP_R2WAFI_MPC__20170206T103047_V20170101T000000_21000101T000000_B08,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B11,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B05,S2B_OPER_GIP_R2WAFI_MPC__20170206T103040_V20170101T000000_21000101T000000_B12,S2B_OPER_GIP_R2WAFI_MPC__20170206T103039_V20170101T000000_21000101T000000_B03,S2B_OPER_GIP_RESPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_SPAMOD_MPC__20250428T000037_V20250430T000000_21000101T000000_B00,S2B_OPER_GIP_TILPAR_MPC__20170206T103032_V20170101T000000_21000101T000000_B00,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B8A,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B01,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B11,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B08,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B03,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B07,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B02,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B12,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B04,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B06,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B09,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B05,S2B_OPER_GIP_VIEDIR_MPC__20170512T114736_V20170322T000000_21000101T000000_B10,S2__OPER_GIP_L2ACSC_MPC__20220121T000003_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_L2ACAC_MPC__20220121T000004_V20220125T022000_21000101T000000_B00,S2__OPER_GIP_PROBA2_MPC__20240809T000511_V20240813T070000_21000101T000000_B00', 'used CAMS file names': 'S2__OPER_AUX_CAMSFO_ADG__20250430T000000_V20250430T000000_20250502T010000', 'used DEM file name': 'CopernicusDEM30', 'used ECMWF file names': 'S2__OPER_AUX_ECMWFD_ADG__20250430T000000_V20250430T090000_20250502T030000', 'used ESA CCI land cover map file name': 'ESACCI-LC-L4-LCCS-Map-300m-P1Y-2015-v2.0.7.tif', 'used ESA CCI snow condition map folder name': 'ESACCI-LC-L4-Snow-Cond-500m-MONTHLY-2000-2012-v2.4', 'used ESA CCI water bodies map file name': 'ESACCI-LC-L4-WB-Map-150m-P13Y-2000-v4.0.tif', 'used GRI file name': 'S2A_OPER_GRI_MSIL1B_MPC__20160521T184422_S20151114T103133,S2A_OPER_GRI_MSIL1B_MPC__20160531T210152_S20160512T102954,S2A_OPER_GRI_MSIL1B_MPC__20160907T095549_S20160422T103554,S2A_OPER_GRI_MSIL1B_MPC__20161018T120000_S20150806T102429,S2A_OPER_GRI_MSIL1B_MPC__20161018T120000_S20150826T102633', 'used IERS file name': 'S2__OPER_AUX_UT1UTC_PDMC_20250424T000000_V20250425T000000_20260424T000000', 'used LibRadTran Look-Up Tables list': 'None', 'used snow climatology map file name': 'GlobalSnowMap.tiff'}, 'organisation': 'ESA', 'output': 'S2MSIL2A_etc', 'processor': 'Sentinel-2 IPF', 'type': 'Level-2A Product', 'version': '???'}], 'horizontal_CRS_code': 'EPSG:32632', 'horizontal_CRS_name': 'WGS84 / UTM zone 32N', 'mean_sensing_time': '2025-04-30T10:28:37.778935Z', 'mean_sun_azimuth_angle_in_deg_for_all_bands_all_detectors': 152.175062917123, 'mean_sun_zenith_angle_in_deg_for_all_bands_all_detectors': 32.3195000325452, 'mean_value_of_aerosol_optical_thickness': 0.128691, 'mean_value_of_total_water_vapour_content': 1.228733, 'meteo': {'source': 'ECMWF', 'type': 'FORECAST'}, 'multispectral_registration_assessment': 'N/A', 'onboard_compression_flag': 'true', 'onboard_equalization_flag': 'null', 'optical_crosstalk_correction_flag': 'null', 'ozone_source': 'AUX_ECMWFT', 'ozone_value': 357.378705, 'percentage_of_degraded_MSI_data': 0.0, 'planimetric_stability_assessment_from_AOCS': '\n ', 'product_quality_status': 'PASSED,PASSED,PASSED,PASSED,PASSED,PASSED', 'reflectance_correction_factor_from_the_Sun-Earth_distance_variation_computed_using_the_acquisition_date': 0.987756572639482, 'spectral_band_of_reference': 'N/A'}
- stac_discovery :
- {'assets': {'analytic': {'eo:bands': [{'center_wavelength': 0.4423, 'common_name': 'coastal', 'full_width_half_max': 0.02, 'name': '01', 'solar_illumination': '1874.3'}, {'center_wavelength': 0.4923, 'common_name': 'blue', 'full_width_half_max': 0.065, 'name': '02', 'solar_illumination': '1959.75'}, {'center_wavelength': 0.559, 'common_name': 'green', 'full_width_half_max': 0.035, 'name': '03', 'solar_illumination': '1824.93'}, {'center_wavelength': 0.665, 'common_name': 'red', 'full_width_half_max': 0.03, 'name': '04', 'solar_illumination': '1512.79'}, {'center_wavelength': 0.7038, 'full_width_half_max': 0.015, 'name': '05', 'solar_illumination': '1425.78'}, {'center_wavelength': 0.7391000000000001, 'full_width_half_max': 0.015, 'name': '06', 'solar_illumination': '1291.13'}, {'center_wavelength': 0.7797000000000001, 'full_width_half_max': 0.02, 'name': '07', 'solar_illumination': '1175.57'}, {'center_wavelength': 0.833, 'common_name': 'nir', 'full_width_half_max': 0.105, 'name': '08', 'solar_illumination': '1041.28'}, {'center_wavelength': 0.864, 'full_width_half_max': 0.02, 'name': '8A', 'solar_illumination': '953.93'}, {'center_wavelength': 0.9432, 'full_width_half_max': 0.02, 'name': '09', 'solar_illumination': '817.58'}, {'center_wavelength': 1.3769, 'common_name': 'cirrus', 'full_width_half_max': 0.03, 'name': '10', 'solar_illumination': '365.41'}, {'center_wavelength': 1.6104, 'common_name': 'swir16', 'full_width_half_max': 0.09, 'name': '11', 'solar_illumination': '247.08'}, {'center_wavelength': 2.1856999999999998, 'common_name': 'swir22', 'full_width_half_max': 0.18, 'name': '12', 'solar_illumination': '87.75'}], 'eo:cloud_cover': 19.450191, 'eo:snow_cover': 6.650147, 'href': 'null'}}, 'bbox': [7.871918704100081, 44.142863623969056, 6.806013472393584, 45.148073182871244], 'geometry': {'coordinates': [[[6.806013472393584, 44.142863623969056], [6.847172984568832, 44.257699806043775], [6.895765992662161, 44.40461676259632], [6.945798496598133, 44.55119099205477], [6.995257370182014, 44.697946240047614], [7.042942343005017, 44.84518269067407], [7.092044923604431, 44.99198102161969], [7.137123557319143, 45.13650884990956], [7.852592618382666, 45.148073182871244], [7.871918704100081, 44.15979663371134], [6.806013472393584, 44.142863623969056]]], 'type': 'Polygon'}, 'id': 'S2B_MSIL2A_20250430T101559_N0511_R065_T32TLQ_20250430T131328.SAFE', 'links': [{'href': './.zattrs.json', 'rel': 'self', 'type': 'application/json'}], 'properties': {'bands': [{'center_wavelength': 442.3, 'common_name': 'coastal', 'full_width_half_max': 0.02, 'name': 'b01', 'solar_illumination': 1874.3}, {'center_wavelength': 492.3, 'common_name': 'blue', 'full_width_half_max': 0.065, 'name': 'b02', 'solar_illumination': 1959.75}, {'center_wavelength': 559.0, 'common_name': 'green', 'full_width_half_max': 0.035, 'name': 'b03', 'solar_illumination': 1824.93}, {'center_wavelength': 665.0, 'common_name': 'red', 'full_width_half_max': 0.03, 'name': 'b04', 'solar_illumination': 1512.79}, {'center_wavelength': 703.8, 'common_name': 'rededge', 'full_width_half_max': 0.015, 'name': 'b05', 'solar_illumination': 1425.78}, {'center_wavelength': 739.1, 'common_name': 'rededge', 'full_width_half_max': 0.015, 'name': 'b06', 'solar_illumination': 1291.13}, {'center_wavelength': 779.7, 'common_name': 'rededge', 'full_width_half_max': 0.02, 'name': 'b07', 'solar_illumination': 1175.57}, {'center_wavelength': 833.0, 'common_name': 'nir', 'full_width_half_max': 0.105, 'name': 'b08', 'solar_illumination': 1041.28}, {'center_wavelength': 864.0, 'common_name': 'nir08', 'full_width_half_max': 0.02, 'name': 'b8a', 'solar_illumination': 953.93}, {'center_wavelength': 943.2, 'common_name': 'nir09', 'full_width_half_max': 0.02, 'name': 'b09', 'solar_illumination': 817.58}, {'center_wavelength': 1376.9, 'common_name': 'cirrus', 'full_width_half_max': 0.03, 'name': 'b10', 'solar_illumination': 365.41}, {'center_wavelength': 1610.4, 'common_name': 'swir16', 'full_width_half_max': 0.09, 'name': 'b11', 'solar_illumination': 247.08}, {'center_wavelength': 2185.7, 'common_name': 'swir22', 'full_width_half_max': 0.18, 'name': 'b12', 'solar_illumination': 87.75}], 'constellation': 'sentinel-2', 'created': '2025-04-30T13:13:28+00:00', 'datetime': None, 'end_datetime': '2025-04-30T10:15:59.024000+00:00', 'eo:cloud_cover': 19.450191, 'eo:snow_cover': 6.650147, 'eopf:baseline': '05.11', 'eopf:data_take_id': 'GS2B_20250430T101559_042562_N05.11', 'eopf:instrument_mode': 'INS-NOBS', 'eopf:resolutions': {'bands 01, 09, 10': '60', 'bands 02, 03, 04, 08': '10', 'bands 05, 06, 07, 8A, 11, 12': '20'}, 'instrument': 'msi', 'mission': 'copernicus', 'platform': 'sentinel-2b', 'processing:expression': 'systematic', 'processing:facility': 'ESA', 'processing:level': 'L2A', 'processing:lineage': 'IPF L2A processor', 'processing:software': {'Sentinel-2 IPF': ' '}, 'processing:version': '', 'product:timeline': 'NRT', 'product:timeliness': 'PT3H', 'product:timeliness_category': 'NRT', 'product:type': 'S02MSIL2A', 'proj:bbox': [300000.0, 4890240.0, 409800.0, 5000040.0], 'proj:epsg': 32632, 'providers': [{'name': 'L2A Processor', 'roles': ['processor']}, {'name': 'ESA', 'roles': ['producer']}], 'sat:absolute_orbit': 42562, 'sat:orbit_state': 'descending', 'sat:platform_international_designator': '2015-028A', 'sat:relative_orbit': 65, 'sci:doi': '10.5270/S2_-znk9xsj', 'start_datetime': '2025-04-30T10:15:59.024000+00:00'}, 'stac_extensions': ['https://stac-extensions.github.io/eopf/v1.0.0/schema.json', 'https://stac-extensions.github.io/eo/v1.1.0/schema.json', 'https://stac-extensions.github.io/sat/v1.0.0/schema.json', 'https://stac-extensions.github.io/view/v1.0.0/schema.json', 'https://stac-extensions.github.io/scientific/v1.0.0/schema.json', 'https://stac-extensions.github.io/processing/v1.2.0/schema.json', 'https://stac-extensions.github.io/product/v0.1.0/schema.json'], 'stac_version': '1.0.0', 'type': 'Feature'}
ds.quality_l2a_quicklook_r60m_tci.plot.imshow(rgb="quality_l2a_quicklook_r60m_band")
<matplotlib.image.AxesImage at 0x330582bd0>
There isn’t just one STAC catalog#
There are many STAC catalogs, each hosting different collections of datasets. A collection groups related assets, such as all Sentinel-2 images for a region, or all Landsat scenes for a certain period. By browsing catalogs and their collections, users can quickly find the data they need without hunting through petabytes of raw files.
The STAC catalog at https://earth-search.aws.element84.com/v1 is provided by Element 84 and offers a centralized search interface for various geospatial datasets hosted on AWS. This catalog is part of the Earth Search project, which is designed to facilitate access to open geospatial data through a standardized SpatioTemporal Asset Catalog (STAC) API.
import stackstac
# West, South, East, North
spatial_extent = [11.1, 46.1, 11.5, 46.5]
temporal_extent = ["2015-01-01","2022-01-01"]
Running this cell may take up to 2 minutes
URL = "https://earth-search.aws.element84.com/v1"
catalog = pystac_client.Client.open(URL)
# Search Sentinel-2 L2A items
items = catalog.search(
bbox=spatial_extent,
datetime=temporal_extent,
collections=["sentinel-2-l2a"]
).item_collection()
Calling stackstac.stack() method for the items, the data will be lazily loaded and an xArray.DataArray object returned.
Running the next cell will show the selected data content with the dimension names and their extent.
# Stack into a datacube
datacube = stackstac.stack(
items,
bounds_latlon=spatial_extent,
epsg=32632, # UTM zone for tile 32N
resolution=10, # 10m pixels
chunksize=4096 # optional, improves performance
)
datacube
<xarray.DataArray 'stackstac-6820bbacf43f782d94d9a1563f88a8ac' (time: 966,
band: 32,
y: 4535, x: 3210)> Size: 4TB
dask.array<fetch_raster_window, shape=(966, 32, 4535, 3210), dtype=float64, chunksize=(1, 1, 4096, 3210), chunktype=numpy.ndarray>
Coordinates: (12/52)
* time (time) datetime64[ns] 8kB 2016-1...
id (time) <U24 93kB 'S2A_32TPS_2016...
* band (band) <U12 2kB 'aot' ... 'wvp-jp2'
* x (x) float64 26kB 6.611e+05 ... 6...
* y (y) float64 36kB 5.153e+06 ... 5...
s2:granule_id (time) <U62 240kB 'S2A_OPER_MSI_...
... ...
raster:bands (band) object 256B [{'nodata': 0...
gsd (band) object 256B None 10 ... None
common_name (band) object 256B None ... None
center_wavelength (band) object 256B None ... None
full_width_half_max (band) object 256B None ... None
epsg int64 8B 32632
Attributes: (4)- time: 966
- band: 32
- y: 4535
- x: 3210
- dask.array<chunksize=(1, 1, 4096, 3210), meta=np.ndarray>
Array Chunk Bytes 3.27 TiB 100.31 MiB Shape (966, 32, 4535, 3210) (1, 1, 4096, 3210) Dask graph 61824 chunks in 3 graph layers Data type float64 numpy.ndarray - time(time)datetime64[ns]2016-11-05T10:12:57.363000 ... 2...
array(['2016-11-05T10:12:57.363000000', '2016-11-08T10:24:25.911000000', '2016-11-15T10:13:01.462000000', ..., '2021-12-29T10:18:00.908000000', '2021-12-29T10:18:00.910000000', '2022-01-01T10:27:57.174000000'], dtype='datetime64[ns]') - id(time)<U24'S2A_32TPS_20161105_0_L2A' ... '...
array(['S2A_32TPS_20161105_0_L2A', 'S2A_32TPS_20161108_0_L2A', 'S2A_32TPS_20161115_0_L2A', 'S2A_32TPS_20161118_0_L2A', 'S2A_32TPS_20161125_0_L2A', 'S2A_32TPS_20161128_0_L2A', 'S2A_32TPS_20161205_0_L2A', 'S2A_32TPS_20161208_0_L2A', 'S2A_32TPS_20161215_0_L2A', 'S2A_32TPS_20161218_0_L2A', 'S2A_32TPS_20161225_0_L2A', 'S2A_32TPS_20161228_0_L2A', 'S2A_32TPS_20170716_0_L2A', 'S2B_32TPS_20180124_0_L2A', 'S2B_32TPS_20180308_0_L2A', 'S2B_32TPS_20180328_0_L2A', 'S2A_32TPS_20180330_0_L2A', 'S2A_32TPS_20180402_0_L2A', 'S2A_32TPS_20180402_1_L2A', 'S2B_32TPS_20180404_0_L2A', 'S2B_32TPS_20180407_0_L2A', 'S2A_32TPS_20180409_0_L2A', 'S2A_32TPS_20180412_0_L2A', 'S2B_32TPS_20180414_0_L2A', 'S2B_32TPS_20180417_0_L2A', 'S2A_32TPS_20180419_0_L2A', 'S2A_32TPS_20180422_0_L2A', 'S2A_32TPS_20180422_1_L2A', 'S2B_32TPS_20180424_0_L2A', 'S2B_32TPS_20180427_0_L2A', 'S2B_32TPS_20180427_1_L2A', 'S2A_32TPS_20180429_0_L2A', 'S2A_32TPS_20180502_0_L2A', 'S2B_32TPS_20180504_0_L2A', 'S2B_32TPS_20180507_0_L2A', 'S2A_32TPS_20180509_0_L2A', 'S2A_32TPS_20180512_0_L2A', 'S2B_32TPS_20180514_0_L2A', 'S2B_32TPS_20180514_1_L2A', 'S2B_32TPS_20180517_0_L2A', ... 'S2B_32TPS_20211028_0_L2A', 'S2A_32TPS_20211030_1_L2A', 'S2A_32TPS_20211030_0_L2A', 'S2A_32TPS_20211102_0_L2A', 'S2A_32TPS_20211102_1_L2A', 'S2B_32TPS_20211104_1_L2A', 'S2B_32TPS_20211104_0_L2A', 'S2B_32TPS_20211107_0_L2A', 'S2A_32TPS_20211112_1_L2A', 'S2B_32TPS_20211114_1_L2A', 'S2B_32TPS_20211114_0_L2A', 'S2B_32TPS_20211117_0_L2A', 'S2A_32TPS_20211119_0_L2A', 'S2A_32TPS_20211122_0_L2A', 'S2B_32TPS_20211124_0_L2A', 'S2B_32TPS_20211127_0_L2A', 'S2A_32TPS_20211129_0_L2A', 'S2A_32TPS_20211202_1_L2A', 'S2A_32TPS_20211202_0_L2A', 'S2B_32TPS_20211204_1_L2A', 'S2B_32TPS_20211204_0_L2A', 'S2B_32TPS_20211207_0_L2A', 'S2A_32TPS_20211209_0_L2A', 'S2A_32TPS_20211212_2_L2A', 'S2A_32TPS_20211212_1_L2A', 'S2A_32TPS_20211212_0_L2A', 'S2B_32TPS_20211214_0_L2A', 'S2B_32TPS_20211217_0_L2A', 'S2B_32TPS_20211217_1_L2A', 'S2A_32TPS_20211219_1_L2A', 'S2A_32TPS_20211219_0_L2A', 'S2A_32TPS_20211222_0_L2A', 'S2B_32TPS_20211224_1_L2A', 'S2B_32TPS_20211224_0_L2A', 'S2B_32TPS_20211227_0_L2A', 'S2A_32TPS_20211229_1_L2A', 'S2A_32TPS_20211229_0_L2A', 'S2A_32TPS_20220101_0_L2A'], dtype='<U24') - band(band)<U12'aot' 'blue' ... 'wvp-jp2'
array(['aot', 'blue', 'coastal', 'green', 'nir', 'nir08', 'nir09', 'red', 'rededge1', 'rededge2', 'rededge3', 'scl', 'swir16', 'swir22', 'visual', 'wvp', 'aot-jp2', 'blue-jp2', 'coastal-jp2', 'green-jp2', 'nir-jp2', 'nir08-jp2', 'nir09-jp2', 'red-jp2', 'rededge1-jp2', 'rededge2-jp2', 'rededge3-jp2', 'scl-jp2', 'swir16-jp2', 'swir22-jp2', 'visual-jp2', 'wvp-jp2'], dtype='<U12') - x(x)float646.611e+05 6.611e+05 ... 6.932e+05
array([661130., 661140., 661150., ..., 693200., 693210., 693220.])
- y(y)float645.153e+06 5.153e+06 ... 5.107e+06
array([5152650., 5152640., 5152630., ..., 5107330., 5107320., 5107310.])
- s2:granule_id(time)<U62'S2A_OPER_MSI_L2A_TL_SHIT_201905...
array(['S2A_OPER_MSI_L2A_TL_SHIT_20190504T200851_A007169_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190504T224516_A007212_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190504T225049_A007312_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190505T012451_A007355_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190505T004539_A007455_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190505T013016_A007498_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190505T201826_A007598_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190505T185759_A007641_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190505T054132_A007741_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190505T143721_A007784_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190505T122049_A007884_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20190508T201139_A007927_T32TPS_N00.01', 'S2A_OPER_MSI_L2A_TL_SHIT_20201008T134008_A010787_T32TPS_N00.01', 'S2B_OPER_MSI_L2A_TL_SHIT_20200930T170455_A004624_T32TPS_N00.01', 'S2B_OPER_MSI_L2A_TL_SHIT_20200315T002708_A005239_T32TPS_N00.01', 'S2B_OPER_MSI_L2A_TL_SGS__20180328T155221_A005525_T32TPS_N02.07', 'S2A_OPER_MSI_L2A_TL_EPAE_20180330T110651_A014462_T32TPS_N02.07', 'S2A_OPER_MSI_L2A_TL_SGS__20180402T155007_A014505_T32TPS_N02.07', 'S2A_OPER_MSI_L2A_TL_S2RP_20230908T074848_A014505_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_EPAE_20180404T113026_A005625_T32TPS_N02.07', ... 'S2B_OPER_MSI_L2A_TL_S2RP_20221223T132023_A024787_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20211204T130603_A024787_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211207T122554_A024830_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS4_20211209T115742_A033767_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_S2RP_20221224T052549_A033810_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20211212T115021_A033810_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211212T131717_A033810_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211214T120410_A024930_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211217T122256_A024973_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_S2RP_20221228T160151_A024973_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20221227T163048_A033910_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20211219T120654_A033910_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211222T131613_A033953_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_S2RP_20221228T143406_A025073_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20211224T123805_A025073_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211227T123108_A025116_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_S2RP_20221227T034720_A034053_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20211229T120455_A034053_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20220101T133200_A034096_T32TPS_N03.01'], dtype='<U62') - eo:cloud_cover(time)object89.802461 41.069562 ... 1.819233
array([89.802461, 41.069562, 21.447816, 83.019969, 98.828253, 11.598649, 10.061224, 3.851569, 7.517994, 2.810787, 39.132269, 4.720581, 2.15216, 1.541246, 19.125673, 47.678897, 83.933474, 3.834101, 6.280085, 93.267879, 16.952058, 99.17289, 99.999997, 8.727837, 30.72951, 2.674829, 4.552688, 4.418409, 20.214072, 77.817195, 85.126668, 86.858769, 99.989082, 94.156861, 70.427212, 81.729768, 77.190344, 98.051484, 99.770707, 85.780076, 99.274296, 99.497696, 77.723987, 76.499369, 86.003697, 49.83463, 58.919817, 95.261597, 85.225672, 71.740605, 77.044225, 61.659343, 64.799374, 88.530826, 88.56613, 97.594714, 99.745363, 10.581071, 46.402426, 52.490646, 31.676527, 7.234149, 24.42363, 25.80952, 97.693309, 98.827285, 23.797498, 30.567166, 46.61635, 52.609819, 78.411372, 83.242798, 19.853057, 22.522238, 50.208156, 13.492065, 52.946688, 53.351909, 15.118092, 19.236591, 98.39745, 56.500712, 39.519855, 37.046246, 13.169393, 17.030643, 58.697453, 61.840159, 18.697968, 23.762882, 53.17258, 56.721526, 59.108125, 68.338931, 61.591433, 69.623345, 38.294726, 45.964068, 19.968619, 25.822678, 31.229971, 38.823864, 46.84215, 54.65501, 76.371446, 88.166429, 86.579043, 90.993309, 3.373786, 2.514534, 80.903928, 93.178124, 96.242404, 26.934207, 28.672209, 43.542319, 7.194714, 10.379995, 26.732709, 23.252083, ... 36.142011, 44.22752, 16.351688, 11.912692, 66.694911, 74.698305, 30.488122, 24.658144, 66.222459, 67.488335, 69.336706, 21.845344, 11.819513, 95.981097, 94.308066, 95.824871, 13.046401, 60.47922, 88.411063, 84.81668, 91.857505, 84.186497, 53.051281, 45.899964, 97.948486, 93.562814, 94.032168, 17.593951, 21.935783, 27.057914, 2.928993, 39.962261, 29.239996, 15.004637, 51.434155, 57.301313, 7.634287, 90.122948, 48.727414, 39.703724, 26.229503, 32.442394, 31.934056, 26.521887, 46.43312, 53.405118, 13.073093, 9.353816, 17.80964, 23.461142, 99.295479, 99.519676, 11.764671, 16.145074, 30.412707, 25.403808, 21.293939, 12.179405, 12.935878, 12.85354, 11.459897, 12.891503, 9.908477, 94.264865, 91.273272, 90.78303, 55.890612, 14.478517, 13.405893, 1.437587, 4.204268, 14.1886, 15.741865, 10.860672, 0.008015, 1.178021, 13.292886, 1.680364, 0.73226, 92.882216, 99.027812, 1.36016, 41.920447, 49.236326, 24.686247, 32.565907, 96.813482, 84.677558, 12.108641, 10.056364, 98.568016, 63.295021, 31.37714, 69.938152, 78.075835, 1.867089, 59.735068, 44.367117, 69.913936, 82.252449, 59.305972, 48.154598, 24.801003, 99.296755, 0.076005, 3.937407, 3.965086, 84.968833, 5.395852, 0.031637, 0.002641, 2.215066, 7.4991, 77.799112, 97.440652, 48.588572, 71.127248, 93.765759, 1.819233], dtype=object) - s2:datatake_type()<U8'INS-NOBS'
array('INS-NOBS', dtype='<U8') - updated(time)<U24'2022-11-08T11:45:48.722Z' ... '...
array(['2022-11-08T11:45:48.722Z', '2022-11-08T12:04:13.289Z', '2022-11-08T11:45:57.228Z', '2022-11-08T11:48:05.334Z', '2022-11-08T11:48:05.102Z', '2022-11-08T11:45:48.030Z', '2022-11-08T11:48:22.923Z', '2022-11-08T11:47:34.155Z', '2022-11-08T12:04:14.741Z', '2022-11-08T11:48:22.390Z', '2022-11-08T11:48:23.433Z', '2022-11-08T11:48:28.986Z', '2022-11-08T11:50:26.339Z', '2022-11-08T11:48:26.509Z', '2022-11-08T12:04:09.409Z', '2022-11-08T11:47:56.010Z', '2022-11-08T11:48:17.112Z', '2022-11-08T11:50:14.096Z', '2024-03-17T06:22:19.457Z', '2022-11-08T11:47:52.653Z', '2022-11-08T11:47:54.685Z', '2022-11-08T12:04:09.418Z', '2022-11-08T12:04:08.507Z', '2022-11-08T11:50:57.906Z', '2022-11-08T11:51:00.574Z', '2022-11-08T11:50:12.616Z', '2022-11-08T12:04:54.762Z', '2024-03-26T02:21:48.168Z', '2022-11-08T12:04:57.795Z', '2022-11-08T11:49:19.981Z', '2024-03-26T01:15:20.524Z', '2022-11-08T11:49:21.312Z', '2022-11-08T11:48:35.657Z', '2022-11-08T11:51:18.657Z', '2022-11-08T11:48:15.815Z', '2022-11-08T11:48:15.796Z', '2022-11-08T12:04:08.733Z', '2022-11-08T12:05:53.592Z', '2024-03-25T19:43:15.001Z', '2022-11-08T12:05:55.072Z', ... '2022-11-06T04:43:01.273Z', '2023-06-26T11:48:45.811Z', '2022-11-06T04:43:12.966Z', '2022-11-06T04:41:58.919Z', '2023-06-24T20:37:58.828Z', '2023-06-30T18:35:20.074Z', '2022-11-06T04:43:58.654Z', '2022-11-06T04:43:51.058Z', '2022-11-06T04:43:59.275Z', '2023-06-21T22:07:52.241Z', '2022-11-06T04:43:09.999Z', '2022-11-06T04:42:57.691Z', '2022-11-06T04:41:58.889Z', '2022-11-06T04:43:35.334Z', '2022-11-06T13:34:46.222Z', '2022-11-06T13:34:45.866Z', '2022-11-06T04:43:39.759Z', '2023-06-21T19:18:07.765Z', '2022-11-06T04:42:35.694Z', '2023-06-21T16:44:11.529Z', '2022-11-06T04:44:51.652Z', '2022-11-06T04:45:13.963Z', '2022-11-06T04:43:06.076Z', '2023-06-21T13:25:26.939Z', '2022-11-06T04:43:49.287Z', '2022-11-06T04:43:54.186Z', '2022-11-06T04:43:31.291Z', '2022-11-06T04:43:13.303Z', '2023-06-29T19:55:36.667Z', '2023-06-30T07:19:57.427Z', '2022-11-06T04:42:57.429Z', '2022-11-06T04:42:53.943Z', '2023-06-29T19:52:20.154Z', '2022-11-06T04:44:49.049Z', '2022-11-06T13:41:28.061Z', '2023-06-30T09:10:08.167Z', '2022-11-06T13:41:29.936Z', '2022-11-06T11:26:42.805Z'], dtype='<U24') - instruments()<U3'msi'
array('msi', dtype='<U3') - earthsearch:s3_path(time)<U80's3://sentinel-cogs/sentinel-s2-...
array(['s3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/11/S2A_32TPS_20161105_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/11/S2A_32TPS_20161108_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/11/S2A_32TPS_20161115_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/11/S2A_32TPS_20161118_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/11/S2A_32TPS_20161125_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/11/S2A_32TPS_20161128_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/12/S2A_32TPS_20161205_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/12/S2A_32TPS_20161208_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/12/S2A_32TPS_20161215_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/12/S2A_32TPS_20161218_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/12/S2A_32TPS_20161225_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2016/12/S2A_32TPS_20161228_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2017/7/S2A_32TPS_20170716_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2018/1/S2B_32TPS_20180124_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2018/3/S2B_32TPS_20180308_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2018/3/S2B_32TPS_20180328_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2018/3/S2A_32TPS_20180330_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2018/4/S2A_32TPS_20180402_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2018/4/S2A_32TPS_20180402_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2018/4/S2B_32TPS_20180404_0_L2A', ... 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211204_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211204_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211207_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211209_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_2_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211214_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211217_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211217_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211219_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211219_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211222_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211224_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211224_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211227_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211229_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211229_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2022/1/S2A_32TPS_20220101_0_L2A'], dtype='<U80') - earthsearch:payload_id(time)<U74'roda-sentinel2/workflow-sentine...
array(['roda-sentinel2/workflow-sentinel2-to-stac/249007ea3ff873c910a70dc02d37037c', 'roda-sentinel2/workflow-sentinel2-to-stac/f399a0fb22b847b69d4d5bfe68397e5e', 'roda-sentinel2/workflow-sentinel2-to-stac/cb2b26e940c89571747a6de3ef8c178c', 'roda-sentinel2/workflow-sentinel2-to-stac/4097fd1aedbe5b76038514477fa127c9', 'roda-sentinel2/workflow-sentinel2-to-stac/a6ed3fdc318adfc0dcdf611834b7a26a', 'roda-sentinel2/workflow-sentinel2-to-stac/cfea8d47e929afa085f7cc819cb3509a', 'roda-sentinel2/workflow-sentinel2-to-stac/3b978306d85aa48d07488b32eaeb0a5c', 'roda-sentinel2/workflow-sentinel2-to-stac/67ecaba97b0043df3f479b522267756c', 'roda-sentinel2/workflow-sentinel2-to-stac/836ade8d7a27e7d1c8126836d6382893', 'roda-sentinel2/workflow-sentinel2-to-stac/cca6df699919ff449cd6f759998038ab', 'roda-sentinel2/workflow-sentinel2-to-stac/308d9bfadeaf73cbc2b86b568ba11cff', 'roda-sentinel2/workflow-sentinel2-to-stac/6c1e6272e2f028763ca41bc4dd16a34a', 'roda-sentinel2/workflow-sentinel2-to-stac/9665a594257d0210dcbbee02641af1e5', 'roda-sentinel2/workflow-sentinel2-to-stac/13086cf32ad4588d24e1e0f9cd93e3e6', 'roda-sentinel2/workflow-sentinel2-to-stac/c8395e7cdb1b9124ca20a0987bd843a7', 'roda-sentinel2/workflow-sentinel2-to-stac/68f29e4e5fd83a2628fe807737219ea0', 'roda-sentinel2/workflow-sentinel2-to-stac/55fe2c964bbb748be34034bf7f6a85e6', 'roda-sentinel2/workflow-sentinel2-to-stac/94f41c1bb92dd28c6049191012a19029', 'roda-sentinel2/workflow-sentinel2-to-stac/12a70d577323012254fd1bdefb25ffbd', 'roda-sentinel2/workflow-sentinel2-to-stac/2ca39f636ff61764181e0ee4c6b36de0', ... 'roda-sentinel2/workflow-sentinel2-to-stac/26e0e95830ac597b1c71d76d22262a86', 'roda-sentinel2/workflow-sentinel2-to-stac/a559c05064075ad2574fcf053cf91401', 'roda-sentinel2/workflow-sentinel2-to-stac/e40555c4205e721d21745274be8f4e0b', 'roda-sentinel2/workflow-sentinel2-to-stac/e926bcb18df86e4c8cc5e11effbd8a54', 'roda-sentinel2/workflow-sentinel2-to-stac/93568ff358f842879ea566c2e766d45d', 'roda-sentinel2/workflow-sentinel2-to-stac/71aba90ac7b3a173c126c68065ea5637', 'roda-sentinel2/workflow-sentinel2-to-stac/8366e35ded0e5bd249b6580f59a52c49', 'roda-sentinel2/workflow-sentinel2-to-stac/70d4667f3fdf5100376fa32008a9997a', 'roda-sentinel2/workflow-sentinel2-to-stac/ad99eb1e478397b6092c8f33bfb2155a', 'roda-sentinel2/workflow-sentinel2-to-stac/62fdb0869502d7847df53e10633177e6', 'roda-sentinel2/workflow-sentinel2-to-stac/a60d1d53197ebe894647143a010a1bb6', 'roda-sentinel2/workflow-sentinel2-to-stac/313a6f02840be6a2663a0e3230e357b2', 'roda-sentinel2/workflow-sentinel2-to-stac/c821731deb57cc090c7762e61fba8553', 'roda-sentinel2/workflow-sentinel2-to-stac/6cfd17cb19fccbcc6ed104c90bea698a', 'roda-sentinel2/workflow-sentinel2-to-stac/b05fc34f20ed5c9182108650c6d58844', 'roda-sentinel2/workflow-sentinel2-to-stac/d62729e79bcc1f5079cc1c14264356f9', 'roda-sentinel2/workflow-sentinel2-to-stac/fe118032f87dd2a9f3eef967eea9450a', 'roda-sentinel2/workflow-sentinel2-to-stac/816b1dd316dc5d71bd3d4e6aeea59e96', 'roda-sentinel2/workflow-sentinel2-to-stac/861880c310ba714513171febecbaef16'], dtype='<U74') - s2:saturated_defective_pixel_percentage()int640
array(0)
- s2:cloud_shadow_percentage(time)object0 3.020158 ... 0.089572 4.863349
array([0, 3.020158, 4.634248, 0.309864, 0.002293, 3.726233, 3.048624, 3.395732, 2.698533, 2.92346, 5.322062, 4.046816, 0.279148, 4.1951, 3.240826, 3.663882, 1.270586, 4.247827, 0.091881, 0.000392, 5.823573, 0, 0, 2.812845, 5.403935, 2.2592, 2.962893, 1.764707, 5.058312, 0.787107, 1.238994, 0.462941, 0, 0.236977, 2.507331, 1.071134, 2.076705, 0.007913, 0.227961, 1.042708, 0.005637, 0, 0.671373, 1.071081, 2.541441, 4.130048, 4.209336, 0.01921, 0.017815, 1.741422, 0.857294, 0.502689, 0.340445, 0.590416, 0.306285, 0, 0.226399, 1.552067, 2.00094, 3.293344, 3.79289, 0.344067, 1.429798, 1.629391, 0.001536, 1.090726, 3.171057, 3.340535, 3.359103, 2.530231, 0.505924, 0.569909, 1.043995, 0.974337, 4.122549, 2.149216, 2.848225, 2.190805, 2.397009, 2.578206, 0.011641, 3.341344, 5.495783, 1.961362, 3.17012, 3.421951, 1.987545, 0.899503, 5.638104, 6.043283, 1.481606, 1.554215, 3.956404, 3.146451, 4.787254, 2.496117, 5.288706, 6.030933, 5.513211, 4.66733, 8.733982, 8.936849, 5.691534, 3.765235, 2.281033, 1.750303, 2.736252, 0.509881, 0.429037, 0.08431, 2.860119, 0.346904, 0.123085, 3.596708, 2.553816, 8.610032, 5.143658, 6.478773, 0.337866, 0.012452, 3.727338, 9.073861, 5.543966, 5.169939, 5.023865, 2.615414, 2.517963, ... 0.556963, 0.533688, 3.479381, 3.173427, 0, 0.013745, 5.264171, 2.58029, 4.672229, 5.893223, 2.496737, 1.637513, 3.359168, 3.461057, 4.921719, 2.398074, 0.05181, 0.063812, 2.271286, 3.426974, 0.44463, 0.045025, 0.056012, 0.025262, 2.726647, 3.151665, 0.262505, 1.31324, 0.048152, 0.491999, 2.331938, 2.918783, 1.875508, 0.109446, 0, 3.420703, 3.360405, 3.427076, 1.115958, 5.910857, 3.698329, 4.988269, 4.066226, 2.495268, 2.338978, 0.255857, 4.353682, 4.655593, 5.854148, 5.005828, 6.813855, 5.979189, 5.98097, 4.044904, 5.527373, 5.125841, 7.878248, 8.526719, 0.519836, 7e-05, 5.936027, 6.944747, 5.906447, 5.701056, 1.727471, 0.01861, 0.911201, 3.208879, 3.736543, 7.158039, 6.067993, 0.077211, 0.558425, 2.78953, 2.938063, 2.205732, 4.499008, 0.815014, 3.411923, 5.359535, 5.697567, 1.869579, 0.001064, 2.300628, 2.528195, 2.946776, 0.841954, 5.98001, 0.004317, 1.919591, 0.105607, 0.821381, 3.19216, 2.183539, 2.335925, 0.655469, 3.649843, 5.309501, 1.211522, 0, 5.149199, 0.779268, 0.008605, 3.069539, 1.229004, 0.906537, 0, 0.001559, 0.249077, 1.4789, 3.607851, 0, 0.080606, 1.95125, 4.199561, 0.248602, 5.022087, 0.07016, 0, 4.618132, 5.073084, 0.135842, 0, 1.621613, 0, 0.089572, 4.863349], dtype=object) - grid:code()<U10'MGRS-32TPS'
array('MGRS-32TPS', dtype='<U10') - processing:software(time)object{'sentinel2-to-stac': '0.1.0'} ....
array([{'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, ... {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}], dtype=object) - s2:processing_baseline(time)<U5'00.01' '00.01' ... '03.01' '03.01'
array(['00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '00.01', '02.07', '02.07', '02.07', '05.00', '02.07', '02.07', '02.07', '02.07', '02.07', '02.07', '02.07', '02.07', '05.00', '02.07', '02.07', '05.00', '02.07', '02.07', '02.07', '02.07', '02.07', '02.07', '02.07', '05.00', '02.07', '02.07', '02.07', '02.08', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '02.08', '05.00', '02.08', '02.08', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '02.08', '02.08', '05.00', '02.08', '05.00', '02.08', '02.08', '02.08', '02.08', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '02.08', '05.00', '05.00', '02.08', '05.00', '02.08', '02.08', '05.00', '02.08', '05.00', '02.08', '02.08', '05.00', '02.08', '05.00', '02.08', '02.08', '02.08', '02.08', '05.00', '02.08', '05.00', '02.08', '05.00', '02.08', '02.08', '02.08', '02.08', '02.09', '02.09', '02.09', '02.09', '02.09', '05.00', '02.09', ... '05.00', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '03.01', '05.00', '05.00', '03.01', '05.00', '03.01', '03.01', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '03.01', '03.01', '03.01', '03.01', '03.01', '03.01', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '05.00', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01', '05.00', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '03.01', '03.01', '03.01', '03.01', '05.00', '03.01', '05.00', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01', '05.00', '03.01', '03.01', '05.00', '03.01', '03.01'], dtype='<U5') - mgrs:grid_square()<U2'PS'
array('PS', dtype='<U2') - platform(time)<U11'sentinel-2a' ... 'sentinel-2a'
array(['sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', ... 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a'], dtype='<U11') - s2:not_vegetated_percentage(time)object0.164157 1.850666 ... 7.659283
array([0.164157, 1.850666, 4.228496, 0.062712, 0.01494, 7.866596, 11.060949, 10.80196, 11.759213, 10.898712, 6.529424, 12.187208, 13.651107, 5.272284, 7.655315, 5.267968, 0.216194, 10.401686, 12.968789, 0.005508, 7.31592, 7e-05, 0, 7.391331, 4.424916, 8.627239, 7.07835, 9.385534, 5.349944, 1.122076, 2.281295, 0.732088, 0.000225, 0.324216, 1.459807, 0.366309, 0.984485, 0.002532, 0.000511, 0.373374, 0.006672, 5.6e-05, 0.817066, 1.631421, 1.530396, 2.411655, 2.718638, 0.430531, 1.55292, 1.590987, 1.523223, 1.619812, 3.706573, 0.481842, 1.368286, 0, 0.028069, 8.395863, 3.896582, 3.698129, 4.001577, 11.015609, 7.485414, 9.71804, 0.026287, 0.049054, 8.355864, 8.719684, 3.643551, 4.550018, 1.156423, 1.332976, 8.364233, 10.362255, 5.091622, 9.503718, 4.097646, 6.245862, 9.042459, 9.856208, 0.013656, 2.53628, 3.846677, 3.862919, 7.077086, 8.180296, 4.495993, 5.406934, 6.582968, 7.424086, 3.547113, 3.627383, 2.384662, 1.720092, 2.943827, 2.398766, 5.27234, 5.134078, 6.961165, 8.391456, 3.755952, 3.793259, 3.145841, 2.92304, 0.542194, 0.55462, 1.362652, 0.702701, 9.698479, 12.791839, 0.576092, 0.112933, 0.425284, 5.035986, 7.563724, 3.338026, 9.456579, 10.631576, 11.160331, 15.452218, 1.831186, 6.442651, ... 4.460402, 3.650089, 8.511797, 9.012622, 2.069628, 1.551549, 7.476269, 8.446728, 3.290667, 2.443062, 2.51325, 7.661092, 11.054097, 0.095346, 1.253951, 0.418496, 10.739637, 3.704729, 0.637141, 0.84085, 0.567433, 0.396651, 3.163189, 4.246458, 0.020995, 0.110404, 0.005855, 7.094216, 8.289152, 6.996013, 11.463607, 4.784627, 6.427905, 8.339526, 2.62776, 2.567404, 12.418957, 0.298508, 3.330533, 2.950784, 6.132471, 6.544104, 5.870067, 6.042008, 4.729842, 4.229522, 8.563243, 8.803445, 6.487073, 5.735015, 0.092584, 0.025713, 11.781086, 15.517755, 6.788647, 3.811718, 11.789299, 17.701979, 13.583587, 17.282544, 11.80613, 13.817489, 10.546423, 1.647448, 0.732975, 1.057581, 0.455294, 8.61038, 3.60538, 12.844381, 8.709615, 10.118182, 5.688804, 8.471844, 16.578038, 11.089849, 12.401794, 13.336709, 19.311985, 0.063002, 0.016821, 15.329108, 12.924264, 6.441978, 1.366863, 5.927436, 0.093029, 0.281522, 3.676585, 5.025407, 0.00137, 0.133689, 2.38993, 2.870575, 0.039828, 7.666632, 0.314422, 0.844861, 1.024943, 0.453029, 4.873255, 0.851776, 2.956193, 0.004025, 6.959495, 0.827069, 2.094016, 0.343366, 6.412929, 10.074805, 10.918631, 6.40268, 7.515902, 0.149847, 0.391017, 2.060978, 2.415762, 0.334853, 7.659283], dtype=object) - proj:code()<U10'EPSG:32632'
array('EPSG:32632', dtype='<U10') - s2:snow_ice_percentage(time)object8.188291 27.784669 ... 26.081771
array([8.188291, 27.784669, 24.088357, 13.318789, 0.750628, 12.01432, 6.756805, 18.257032, 7.929872, 15.523706, 14.1028, 13.105176, 2.482953, 41.874111, 38.054755, 29.105544, 10.980279, 46.408647, 49.294683, 6.507387, 42.403331, 0.779639, 0, 44.81712, 33.760557, 40.94322, 37.729022, 40.871131, 29.619467, 13.461216, 7.222129, 2.874207, 9.4e-05, 1.525678, 13.301969, 8.152296, 3.061996, 0.711889, 0, 1.888655, 0.325716, 0.013931, 10.286286, 6.698538, 1.598508, 6.226337, 2.385288, 1.169615, 7.447045, 3.341286, 1.564481, 2.105594, 1.147678, 1.38384, 1.147714, 2.370868, 0.000169, 7.181017, 2.263783, 1.766145, 4.466031, 5.112977, 3.735559, 4.477243, 1.610084, 0.002057, 3.101788, 2.707154, 2.863852, 1.344004, 3.268357, 0.542084, 1.885674, 1.977386, 2.525691, 2.691312, 2.139489, 1.802046, 2.010621, 2.335911, 0.925806, 0.336331, 0.853096, 1.786002, 1.566542, 1.549209, 0.796325, 0.512148, 1.413358, 1.127203, 0.373249, 0.287428, 1.964584, 0.03832, 1.207351, 0.087013, 1.053642, 1.074078, 1.079661, 1.117926, 0.994811, 0.349802, 0.943333, 0.159254, 8.298742, 0.487451, 0.139014, 0.020036, 3.070256, 2.750362, 5.31062, 2.241406, 0.026867, 2.569779, 2.207145, 2.843166, 2.103161, 2.245224, 1.899111, 2.143112, 1.672328, 1.110504, ... 3.306192, 1.222734, 5.971161, 6.136174, 3.488879, 0.45746, 5.297454, 5.166665, 1.32077, 1.361042, 0.438592, 3.917798, 4.367699, 1.7435, 0.284538, 0.114412, 2.55006, 3.005265, 0.093092, 0.442713, 0.007429, 6.16691, 0.665539, 1.712255, 5.3e-05, 4.084728, 5.961398, 2.40996, 2.481955, 0.888083, 1.795862, 2.171722, 2.065423, 1.489332, 1.351889, 0.182908, 1.572138, 2.445913, 1.516561, 4.81597, 1.950167, 1.802839, 1.449619, 2.754777, 1.69837, 1.075136, 1.543902, 1.469219, 1.413827, 0.885632, 0.000139, 0.037654, 1.82899, 2.053182, 6.347398, 7.540168, 2.476955, 2.796105, 2.337531, 1.900938, 1.758422, 2.897349, 2.63615, 0.229127, 2.455568, 0.078132, 32.46749, 17.610456, 17.242524, 10.428437, 7.091185, 12.082797, 9.418903, 5.790321, 8.214249, 6.919226, 5.038564, 4.362707, 5.516725, 3.6e-05, 0, 3.702908, 2.676089, 0.574998, 31.396148, 25.521684, 0.012097, 10.066652, 25.313136, 27.462873, 0.000979, 35.982648, 27.479434, 8.393566, 20.561327, 20.145309, 28.063837, 40.528587, 28.309372, 12.937294, 25.167498, 34.66765, 31.567097, 0.305201, 54.500091, 49.163428, 42.097259, 5.702758, 29.398009, 39.025801, 36.363328, 31.210962, 23.738836, 21.839589, 0.12061, 22.458659, 24.725914, 2.558568, 26.081771], dtype=object) - s2:product_uri(time)<U65'S2A_MSIL2A_20161105T101212_N000...
array(['S2A_MSIL2A_20161105T101212_N0001_R022_T32TPS_20190504T200850.SAFE', 'S2A_MSIL2A_20161108T102232_N0001_R065_T32TPS_20190504T224514.SAFE', 'S2A_MSIL2A_20161115T101302_N0001_R022_T32TPS_20190504T225048.SAFE', 'S2A_MSIL2A_20161118T102322_N0001_R065_T32TPS_20190505T012450.SAFE', 'S2A_MSIL2A_20161125T101342_N0001_R022_T32TPS_20190505T004538.SAFE', 'S2A_MSIL2A_20161128T102352_N0001_R065_T32TPS_20190505T013015.SAFE', 'S2A_MSIL2A_20161205T101402_N0001_R022_T32TPS_20190505T201824.SAFE', 'S2A_MSIL2A_20161208T102422_N0001_R065_T32TPS_20190505T185758.SAFE', 'S2A_MSIL2A_20161215T101422_N0001_R022_T32TPS_20190505T054130.SAFE', 'S2A_MSIL2A_20161218T102432_N0001_R065_T32TPS_20190505T143719.SAFE', 'S2A_MSIL2A_20161225T101432_N0001_R022_T32TPS_20190505T122048.SAFE', 'S2A_MSIL2A_20161228T102432_N0001_R065_T32TPS_20190508T201138.SAFE', 'S2A_MSIL2A_20170716T102021_N0001_R065_T32TPS_20201008T134006.SAFE', 'S2B_MSIL2A_20180124T101309_N0001_R022_T32TPS_20200930T170453.SAFE', 'S2B_MSIL2A_20180308T102019_N0001_R065_T32TPS_20200315T002706.SAFE', 'S2B_MSIL2A_20180328T102019_N0207_R065_T32TPS_20180328T155221.SAFE', 'S2A_MSIL2A_20180330T101021_N0207_R022_T32TPS_20180330T110651.SAFE', 'S2A_MSIL2A_20180402T102021_N0207_R065_T32TPS_20180402T155007.SAFE', 'S2A_MSIL2A_20180402T102021_N0500_R065_T32TPS_20230908T074848.SAFE', 'S2B_MSIL2A_20180404T101019_N0207_R022_T32TPS_20180404T113026.SAFE', ... 'S2B_MSIL2A_20211204T101309_N0500_R022_T32TPS_20221223T132023.SAFE', 'S2B_MSIL2A_20211204T101309_N0301_R022_T32TPS_20211204T130603.SAFE', 'S2B_MSIL2A_20211207T102319_N0301_R065_T32TPS_20211207T122554.SAFE', 'S2A_MSIL2A_20211209T101411_N0301_R022_T32TPS_20211209T115742.SAFE', 'S2A_MSIL2A_20211212T102431_N0500_R065_T32TPS_20221224T052549.SAFE', 'S2A_MSIL2A_20211212T102431_N0301_R065_T32TPS_20211212T115021.SAFE', 'S2A_MSIL2A_20211212T102431_N0301_R065_T32TPS_20211212T131717.SAFE', 'S2B_MSIL2A_20211214T101329_N0301_R022_T32TPS_20211214T120410.SAFE', 'S2B_MSIL2A_20211217T102329_N0301_R065_T32TPS_20211217T122256.SAFE', 'S2B_MSIL2A_20211217T102329_N0500_R065_T32TPS_20221228T160151.SAFE', 'S2A_MSIL2A_20211219T101431_N0500_R022_T32TPS_20221227T163048.SAFE', 'S2A_MSIL2A_20211219T101431_N0301_R022_T32TPS_20211219T120654.SAFE', 'S2A_MSIL2A_20211222T102441_N0301_R065_T32TPS_20211222T131613.SAFE', 'S2B_MSIL2A_20211224T101329_N0500_R022_T32TPS_20221228T143406.SAFE', 'S2B_MSIL2A_20211224T101329_N0301_R022_T32TPS_20211224T123805.SAFE', 'S2B_MSIL2A_20211227T102339_N0301_R065_T32TPS_20211227T123108.SAFE', 'S2A_MSIL2A_20211229T101431_N0500_R022_T32TPS_20221227T034720.SAFE', 'S2A_MSIL2A_20211229T101431_N0301_R022_T32TPS_20211229T120455.SAFE', 'S2A_MSIL2A_20220101T102431_N0301_R065_T32TPS_20220101T133200.SAFE'], dtype='<U65') - s2:degraded_msi_data_percentage(time)object0 0 0 0 0 0 ... 0 0 0.0038 0 0
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.1174, 0, 0, 0, 0, 0, 0, 0, 0, 0.0259, 0, 0, 0.0278, 0, 0, 0, 0, 0, 0, 0, 0.0346, 0, 0, 0, 0, 0, 0.1774, 0, 0.2152, 0, 0.0114, 0, 0.1139, 0, 0.0426, 0, 1.3, 0, 0.0107, 0, 0, 0.0962, 0, 0, 0, 0.0201, 0, 0.035, 0, 0.0298, 0, 0.3337, 0, 0.0608, 0, 0.0321, 0, 0, 0, 0.0333, 0, 0.0317, 0, 0, 0, 0, 0, 0.0343, 0, 0.0225, 0, 0.0342, 0, 0.0339, 0, 0.1181, 0, 0.065, 0, 0.0186, 0, 0.0361, 0, 0.0695, 0, 0.1018, 0, 0, 0.0248, 0.0137, 0, 0.0312, 0, 0, 0.0451, 0, 0.0132, 0, 0, 0.0257, 0, 0.0109, 0, 0, 0, 0, 0.011, 0, 0.0129, 0, 0.0315, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.0249, 0, 0, 0, 0, 0, 0, 0.0127, 0, 0, 0, 0, 0, 0, 0.0248, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.0107, 0, 0, 0, 0, 0.0107, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.0129, 0, 0, 0, 0, 0, 0, 0, 0, 0.0129, 0, 0.0358, 0, 0.0263, 0, 0.0111, 0, 0.0138, 0, 0, 0.072, 0, 0.0134, 0, 0.1122, 0, 0.0505, 0, 0.0554, 0, 0.0728, 0, 0.013, 0, 0.0854, 0, 0, 0.2268, 0, 0.1952, 0, 0.0544, 0.3582, 0, 0.0297, 0, 0.5618, 0, 0.0308, 0, 0.3514, 0, 0.0569, 0, 0.0265, 0, 0.0365, 0, 0, 0.0246, 0.0149, 0, 0, 0.0148, 0.0246, 0, 0, 0.3598, 0, 0.3168, 0.0056, 0, 0, 0.1237, 0.166, 0, 0, 0.299, 0.0276, 0, 0.0257, 0, 0.0111, 0, 0, 0.2563, 2.0973, 0, 0.0262, 0, 0.2713, 0, 0, 0.0126, 0.0096, 0, 0, 0.0259, ... 0, 0, 0.0253, 0.0107, 0, 0.0222, 0, 0, 0.0035, 0.0246, 0, 0, 0, 0.0223, 0.0035, 0, 0.0106, 0, 0, 0.0226, 0, 0.0252, 0.0105, 0, 0, 0.023, 0, 0.0248, 0, 0, 0.0109, 0, 0.023, 0.0051, 0.0051, 0, 0.025, 0, 0.0109, 0, 0.0224, 0, 0.0036, 0, 0, 0.0248, 0.0108, 0, 0, 0.0231, 0.0035, 0, 0, 0.0253, 0, 0, 0, 0, 0.0318, 0, 0, 0.0437, 0.0051, 0, 0.026, 0, 0, 0, 0.0045, 0, 0.0552, 0, 0.0362, 0, 0.0212, 0, 0.0236, 0, 0, 0, 0.2247, 0.1128, 0.1128, 0, 0.1107, 0, 0.0063, 0, 0.0611, 0, 0, 0.1653, 0, 0, 0.0325, 0, 0.018, 0, 0.0887, 0, 0.1658, 0, 0, 0.0277, 0.2655, 0, 0, 0.0383, 0, 0.0996, 0, 0.0112, 0, 0.0265, 0, 0.1008, 0, 0, 0.2234, 0.1724, 0, 0.0443, 0, 0.0044, 0, 0, 0.0258, 0.0333, 0, 0, 0.0306, 0.0309, 0, 0, 0.1149, 0.0127, 0, 0, 0.6361, 0.0507, 0, 0.0251, 0, 0, 0, 0, 0, 0.1389, 0, 0, 0, 0.0943, 0, 0.5272, 0, 0.0459, 0, 1.4163, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.0263, 0, 0, 0.0215, 0, 0, 0.0268, 0.056, 0, 0, 0.027, 0.0059, 0, 0, 0.0259, 0.0106, 0, 0, 0.0227, 0.0047, 0, 0, 0.0108, 0, 0.0228, 0, 0.0039, 0, 0.0246, 0, 0.0135, 0, 0.0231, 0, 0.0037, 0, 0.0255, 0, 0, 0.0231, 0, 0, 0, 0.0248, 0.0106, 0, 0, 0.0036, 0, 0, 0.0255, 0.0106, 0, 0, 0, 0.0106, 0, 0, 0, 0, 0, 0, 0, 0.0245, 0, 0.0106, 0, 0, 0, 0.0212, 0, 0, 0, 0, 0.0223, 0.0039, 0, 0, 0.0106, 0, 0, 0.0038, 0, 0], dtype=object) - s2:dark_features_percentage(time)object1.844513 14.630641 ... 33.430809
array([1.844513, 14.630641, 19.814493, 2.271957, 0.318426, 31.197238, 34.837586, 33.818662, 35.73682, 35.501686, 18.373249, 8.437745, 1.94334, 28.051034, 16.518265, 3.76165, 0.679082, 6.121882, 12.049133, 0.007064, 6.423968, 0.000279, 0, 2.896829, 3.136145, 3.277949, 3.874581, 7.278542, 2.434123, 0.620643, 0.649142, 0.259382, 0, 0.136526, 0.798275, 0.253184, 0.499691, 0.006503, 0.000159, 0.2124, 0.000962, 0, 0.196546, 0.347806, 0.083968, 0.882459, 0.812422, 0.03581, 0.039983, 0.621944, 1.101459, 0.269276, 0.34078, 0.153071, 0.344264, 0, 0, 1.433662, 0.828302, 0.436869, 1.336161, 0.676073, 1.799984, 2.075599, 0.000458, 0, 2.255403, 2.266792, 0.721617, 1.014303, 0.308301, 0.287663, 1.23983, 1.487523, 1.984327, 1.540386, 1.814582, 2.42098, 1.881459, 2.147481, 0.007626, 0.685601, 1.893222, 0.929015, 2.942146, 3.360493, 0.829855, 1.496837, 2.7531, 3.350742, 0.66581, 0.963524, 0.861513, 1.343914, 1.165383, 1.944794, 3.351847, 3.510491, 2.531372, 3.797224, 2.630224, 3.119748, 1.360481, 2.110047, 0.594295, 0.423773, 0.521823, 1.03964, 2.213023, 3.817084, 0.710635, 0.066938, 0.154753, 3.27911, 4.891308, 2.631714, 5.024238, 6.376091, 4.311187, 6.832167, 2.215226, 4.492992, 5.564722, 6.49466, 4.853905, 5.835064, 4.648668, 6.326275, ... 0.00291, 0.481124, 0.970329, 1.271975, 0.97468, 0.692506, 0.941477, 0.680673, 0.62224, 0.627178, 1.342292, 0.341817, 0.230835, 0.413711, 1.556587, 0.45399, 0.006646, 0.017346, 0.018288, 1.702618, 1.080972, 0.449545, 0.144803, 0.32416, 0.14557, 1.350529, 0.897261, 0.005332, 0.167664, 0.000441, 1.997804, 2.577513, 1.417756, 2.958946, 1.362398, 2.818952, 2.11347, 1.295276, 2.47188, 2.909188, 0.179586, 2.370656, 1.528137, 3.311956, 4.68837, 3.621467, 3.054734, 2.492234, 3.612776, 4.941226, 3.938032, 4.406236, 5.136371, 0.007498, 0.001828, 6.376003, 3.453626, 2.293115, 2.924517, 6.151615, 7.052542, 6.861491, 6.710172, 8.241988, 6.533402, 7.349457, 0.547123, 0.803161, 0.441412, 1.366814, 8.667428, 7.683007, 11.972662, 8.467923, 9.474287, 8.471502, 9.115031, 14.991429, 10.522097, 14.116709, 13.383521, 18.600589, 1.045308, 0.07148, 15.634122, 12.686016, 10.417864, 10.231411, 11.911369, 0.637589, 3.045999, 22.90576, 23.055506, 0.215819, 0.568309, 15.369792, 10.488391, 0.486217, 30.60174, 6.680006, 7.107484, 0.588372, 3.797914, 7.24856, 8.510101, 20.064506, 0.392325, 35.566777, 29.157805, 30.762979, 7.485075, 32.97202, 40.977016, 42.140675, 33.06075, 33.41836, 0.074081, 2.040348, 17.591031, 1.188085, 2.166522, 33.430809], dtype=object) - s2:vegetation_percentage(time)object0 5.312433 ... 0.186854 15.447186
array([0, 5.312433, 18.813984, 0.014458, 0, 24.356632, 24.984224, 20.961724, 24.303922, 21.553582, 9.813931, 21.272539, 77.316386, 9.904308, 5.794503, 1.832371, 0.516106, 21.430971, 18.805209, 8.3e-05, 13.229795, 0, 0, 23.690403, 16.340066, 36.04008, 37.290215, 35.563955, 30.829772, 2.126506, 1.944584, 5.88359, 0, 1.50256, 7.884338, 4.888298, 12.676103, 0.010697, 0, 7.938007, 0.092853, 0, 6.241169, 9.256854, 7.010388, 31.973439, 29.630259, 2.013404, 2.55116, 16.347454, 15.121748, 29.298761, 27.528173, 6.416893, 6.272815, 0, 0, 67.18092, 40.518841, 37.153655, 50.044495, 72.344142, 56.867987, 54.324675, 0.126386, 0.019781, 54.267043, 50.856161, 37.392727, 35.541752, 14.283578, 13.06064, 63.095343, 61.019069, 29.831371, 65.956104, 30.818903, 29.960394, 65.222985, 62.599689, 0.114959, 31.561637, 43.681267, 50.20746, 67.310435, 64.919502, 25.642568, 25.081265, 59.485084, 56.606835, 37.243804, 35.330725, 24.752368, 22.606358, 22.62131, 20.967855, 40.195802, 36.287114, 57.232463, 54.153079, 46.884814, 42.541674, 34.930125, 32.95739, 6.405883, 4.393529, 5.72221, 4.189468, 77.577263, 77.220213, 4.526906, 2.63054, 2.400365, 52.899241, 51.575267, 31.988549, 66.34708, 62.885863, 51.224393, 51.601136, 17.243898, 38.383076, 55.80973, 63.795406, 61.335045, 66.239333, ... 1.309192, 1.238064, 51.928717, 56.201601, 46.205747, 42.820308, 65.318006, 67.6287, 20.386443, 18.003245, 50.318938, 54.387307, 24.66324, 24.224809, 21.713674, 57.629281, 68.127602, 1.215864, 2.659394, 2.681026, 64.335382, 21.454737, 8.762148, 9.374209, 5.740051, 6.012767, 34.98106, 37.54867, 0.127037, 0.799231, 0, 63.6527, 56.566775, 55.664939, 76.903182, 38.662574, 50.731623, 62.95644, 32.21547, 31.252652, 69.595343, 4.149215, 36.914611, 40.034437, 51.235604, 47.250777, 48.697516, 51.247597, 30.600074, 29.438981, 65.299129, 68.121457, 57.551217, 54.01482, 0.06853, 0.100915, 57.554764, 54.942769, 45.802, 48.965672, 52.649057, 59.904116, 60.774791, 56.98514, 58.596641, 55.844522, 58.502585, 0.979335, 0.930941, 2.99717, 3.092943, 46.137369, 47.877368, 62.104517, 63.009542, 47.673592, 49.717781, 58.927053, 59.844887, 61.083966, 47.538799, 56.53919, 54.613954, 0.007724, 0.04288, 53.221428, 27.392629, 28.892845, 22.205168, 21.007425, 0.087744, 0.373337, 25.320077, 21.666245, 0, 0, 9.545267, 3.574096, 0.000522, 27.581957, 0.321148, 0.989128, 0.000337, 0.000961, 0.985843, 1.207169, 7.617316, 0, 2.711944, 3.611166, 5.17522, 0.026513, 10.205209, 9.482228, 10.255733, 11.158184, 11.594474, 0, 0, 1.603692, 0.169973, 0.186854, 15.447186], dtype=object) - s2:thin_cirrus_percentage(time)object0.682698 2.235039 ... 0.176584
array([0.682698, 2.235039, 3.950845, 0.051749, 0.40537, 0.08172, 0.003162, 3.047128, 7e-06, 0.760679, 10.637713, 1.882233, 0.012506, 0.518369, 0.214922, 11.69931, 0.197763, 0.000314, 0.633787, 0.036314, 0.002874, 0.211917, 0, 3.034208, 0.282217, 5.6e-05, 0.028343, 0.016487, 0.33055, 3.226072, 2.60025, 9.320878, 4.9e-05, 2.231814, 4.152849, 0.419537, 0.526925, 0.005292, 2.056553, 2.109121, 2.245188, 0.000348, 0.345225, 0.448363, 0.628233, 0.933827, 1.188463, 2.854118, 2.869688, 7.738028, 7.284503, 3.850004, 4.653349, 10.837879, 10.197846, 0.19913, 0.536061, 0.06716, 0.356386, 1.245185, 0.276729, 0.76527, 4.523561, 5.501174, 0.103215, 0.099704, 0.228731, 0.202629, 1.95438, 2.529219, 0.631788, 1.609778, 1.723737, 2.654188, 0.40214, 0.02789, 12.325025, 11.763097, 0.537677, 0.408599, 1.146258, 0.081593, 0.432907, 0.695771, 0.016387, 0.016005, 2.637755, 2.379843, 0.197689, 0.151274, 0.834014, 2.057893, 0.328009, 1.865363, 0.360399, 0.916435, 0.557557, 1.162105, 0.565177, 1.054937, 0.746099, 2.061865, 1.361463, 2.445826, 3.120027, 1.717414, 3.761454, 1.748552, 3.6e-05, 0.001486, 0.7607, 0.353191, 1.121952, 6.145518, 6.025714, 1.259575, 0.73929, 1.912636, 20.738943, 20.43535, 1.600698, 0.343539, 1.994864, 0.273407, 0.765951, ... 0.684867, 0.146924, 0.19993, 0.375375, 0.267541, 0.697274, 0.426833, 1.096617, 1.318619, 0.212023, 0.239784, 4.645241, 4.688711, 0.15916, 0.133151, 2.168762, 1.993072, 2.624278, 2.716381, 0.519817, 1.241157, 2.319692, 1.521488, 3.397198, 3.406134, 4.356008, 3.906189, 0.782064, 0.357869, 0.005433, 0.085385, 1.93506, 0.155972, 0.002119, 0.197674, 0.018513, 0.058089, 0.076523, 0.709624, 0.426969, 4.070798, 2.62246, 1.025826, 5.238079, 6.772781, 0.442045, 0.237037, 0.10682, 0.20348, 0.387404, 0.06718, 0.264021, 0.750315, 0.201691, 0.21556, 0.027219, 0.011403, 2.562938, 1.423652, 7.697338, 10.991221, 11.106932, 6.085526, 6.020249, 0.030975, 0.009821, 0.522244, 0.603828, 7.088195, 0.155195, 5.873664, 4.157568, 0.003417, 0.001377, 0.994671, 1.122608, 1.056448, 0.000124, 8e-06, 8.33111, 0.000173, 0.010281, 1.691073, 1.701965, 1.1e-05, 6.680907, 6.752215, 2.259765, 3.152109, 1.203165, 1.16127, 0.133605, 5.665855, 3.195145, 0.956709, 0.177512, 19.328448, 1.070345, 0.056168, 0.247231, 8.623155, 2.142848, 2.242485, 9.271787, 9.079917, 1.029431, 0.196121, 0, 0.001398, 0.010097, 0.716185, 1.1e-05, 3.7e-05, 0.000179, 0.000534, 0.042559, 0.296582, 0.330878, 0.779113, 3.44329, 3.387205, 0.176584], dtype=object) - s2:generation_time(time)<U27'2019-05-04T20:08:50.339Z' ... '...
array(['2019-05-04T20:08:50.339Z', '2019-05-04T22:45:14.389Z', '2019-05-04T22:50:48.946Z', '2019-05-05T01:24:50.302Z', '2019-05-05T00:45:38.567Z', '2019-05-05T01:30:15.622Z', '2019-05-05T20:18:24.840Z', '2019-05-05T18:57:58.223Z', '2019-05-05T05:41:30.679Z', '2019-05-05T14:37:19.822Z', '2019-05-05T12:20:48.311Z', '2019-05-08T20:11:38.477Z', '2020-10-08T13:40:06.266Z', '2020-09-30T17:04:53.517Z', '2020-03-15T00:27:06.419Z', '2018-03-28T17:52:18Z', '2018-03-30T11:36:19Z', '2018-04-02T17:17:14Z', '2023-09-08T07:48:48.000000Z', '2018-04-04T12:49:25Z', '2018-04-07T15:23:33Z', '2018-04-09T17:48:39Z', '2018-04-12T13:28:14Z', '2018-04-14T13:33:49Z', '2018-04-17T17:17:27Z', '2018-04-19T11:56:06Z', '2018-04-22T15:38:28Z', '2023-09-15T16:19:12.000000Z', '2018-04-24T13:08:01Z', '2018-04-27T14:39:35Z', '2023-09-13T23:40:11.000000Z', '2018-04-29T11:35:46Z', '2018-05-02T16:32:33Z', '2018-05-04T14:31:48Z', '2018-05-07T13:17:56Z', '2018-05-09T16:21:49Z', '2018-05-12T15:14:13Z', '2018-05-14T15:14:43Z', '2023-09-06T04:01:41.000000Z', '2018-05-17T13:36:20Z', ... '2021-10-28T12:19:42.000000Z', '2023-01-11T21:46:48.000000Z', '2021-10-30T12:05:10.000000Z', '2021-11-02T13:31:55.000000Z', '2023-01-12T18:04:08.000000Z', '2023-01-01T08:28:34.000000Z', '2021-11-04T13:08:49.000000Z', '2021-11-07T12:32:50.000000Z', '2021-11-12T13:19:01.000000Z', '2023-01-01T02:21:49.000000Z', '2021-11-14T12:09:13.000000Z', '2021-11-17T12:31:08.000000Z', '2021-11-19T12:06:04.000000Z', '2021-11-22T13:15:54.000000Z', '2021-11-24T12:26:23.000000Z', '2021-11-27T12:28:14.000000Z', '2021-11-29T11:58:22.000000Z', '2022-12-24T04:54:20.000000Z', '2021-12-02T13:32:27.000000Z', '2022-12-23T13:20:23.000000Z', '2021-12-04T13:06:03.000000Z', '2021-12-07T12:25:54.000000Z', '2021-12-09T11:57:42.000000Z', '2022-12-24T05:25:49.000000Z', '2021-12-12T11:50:21.000000Z', '2021-12-12T13:17:17.000000Z', '2021-12-14T12:04:10.000000Z', '2021-12-17T12:22:56.000000Z', '2022-12-28T16:01:51.000000Z', '2022-12-27T16:30:48.000000Z', '2021-12-19T12:06:54.000000Z', '2021-12-22T13:16:13.000000Z', '2022-12-28T14:34:06.000000Z', '2021-12-24T12:38:05.000000Z', '2021-12-27T12:31:08.000000Z', '2022-12-27T03:47:20.000000Z', '2021-12-29T12:04:55.000000Z', '2022-01-01T13:32:00.000000Z'], dtype='<U27') - constellation()<U10'sentinel-2'
array('sentinel-2', dtype='<U10') - s2:product_type()<U7'S2MSI2A'
array('S2MSI2A', dtype='<U7') - s2:reflectance_conversion_factor(time)float641.016 1.018 1.021 ... 1.034 1.034
array([1.01622743, 1.01776269, 1.02115259, 1.02252252, 1.02547509, 1.02663539, 1.0290544 , 1.02996701, 1.03177239, 1.03240681, 1.03353809, 1.03387299, 0.96766846, 1.03275145, 1.01717327, 1.00626263, 1.00512256, 1.00339524, 1.00339524, 1.00224751, 1.00051506, 0.99936811, 0.99764344, 0.99650591, 0.99480151, 0.99368148, 0.99200973, 0.99200973, 0.99091541, 0.98928829, 0.98928829, 0.98822727, 0.98665612, 0.98563598, 0.98413183, 0.98315947, 0.98173254, 0.98081475, 0.98081475, 0.97947484, 0.97861766, 0.97737366, 0.97658301, 0.97472412, 0.97472412, 0.97369611, 0.97369611, 0.9730534 , 0.9730534 , 0.97214378, 0.97214378, 0.97158158, 0.97158158, 0.97079627, 0.97079627, 0.97031822, 0.97031822, 0.96966227, 0.96927142, 0.96927142, 0.96874902, 0.96844785, 0.96806233, 0.96806233, 0.96785276, 0.96785276, 0.96760654, 0.96760654, 0.96748991, 0.96748991, 0.96738454, 0.96738454, 0.96736157, 0.96736157, 0.96739772, 0.96746857, 0.967646 , 0.967646 , 0.96781022, 0.96781022, 0.96812782, 0.96838435, 0.96884013, 0.96918738, 0.96977843, 0.96977843, 0.97021414, 0.97021414, 0.97093672, 0.97093672, 0.97145816, 0.97145816, 0.97230763, 0.97230763, 0.97291142, 0.97291142, 0.9738823 , 0.9738823 , 0.97456457, 0.97456457, ... 0.9695916 , 0.97001214, 0.97001214, 0.97071125, 0.97121659, 0.97204341, 0.9726335 , 0.97358286, 0.97425016, 0.97531576, 0.97606015, 0.97723516, 0.97723516, 0.97804701, 0.9793236 , 0.98020275, 0.98020275, 0.98157237, 0.98157237, 0.98250735, 0.98250735, 0.98396087, 0.98396087, 0.98495124, 0.98495124, 0.98647864, 0.98647864, 0.98751164, 0.98751164, 0.98910253, 0.98910253, 0.99017676, 0.99017676, 0.99181993, 0.9929235 , 0.9929235 , 0.99460618, 0.99460618, 0.99573409, 0.99573409, 0.99744591, 0.99744591, 0.9985881 , 0.9985881 , 1.00031599, 1.00031599, 1.00146456, 1.00146456, 1.00319682, 1.00319682, 1.00434323, 1.0060657 , 1.0060657 , 1.00720174, 1.00890228, 1.00890228, 1.01001856, 1.01001856, 1.01168368, 1.01277283, 1.01277283, 1.01438977, 1.01438977, 1.01544203, 1.01544203, 1.01699809, 1.01948942, 1.02044477, 1.02044477, 1.02184259, 1.02273838, 1.02403981, 1.02486761, 1.02606217, 1.02681624, 1.02789398, 1.02789398, 1.0285672 , 1.0285672 , 1.02951928, 1.03010686, 1.03092533, 1.03092533, 1.03092533, 1.03142161, 1.03209956, 1.03209956, 1.03250105, 1.03250105, 1.0330328 , 1.03333552, 1.03333552, 1.03371658, 1.03391841, 1.03391841, 1.0341455 ]) - s2:medium_proba_clouds_percentage(time)object64.458448 16.624644 ... 0.527611
array([64.458448, 16.624644, 10.629322, 3.622422, 3.478495, 8.348283, 9.31251, 0.390948, 6.343114, 0.879259, 4.337768, 1.463836, 0.802488, 0.655386, 18.340707, 9.27588, 5.350894, 2.060234, 4.412005, 5.253417, 3.547948, 22.093184, 66.329372, 4.203285, 4.104701, 2.155776, 1.703789, 2.196538, 3.767255, 5.740421, 14.366813, 9.935989, 63.188833, 18.121098, 18.515423, 14.80456, 4.919985, 13.403758, 33.028662, 12.834114, 31.238037, 48.551035, 16.813529, 6.413785, 16.036765, 6.802138, 16.052623, 60.705054, 50.952244, 11.306873, 17.106836, 11.8199, 14.958666, 37.666965, 38.450885, 94.186819, 96.156698, 2.246625, 5.163473, 10.756102, 4.568418, 2.513156, 6.498498, 8.035301, 48.740557, 50.021744, 4.439171, 11.930807, 13.485529, 19.433503, 60.86005, 64.806503, 5.238049, 7.869184, 6.481417, 3.254558, 18.499894, 19.555344, 3.719354, 8.438505, 16.095945, 5.761161, 5.034408, 5.575592, 2.971979, 7.114807, 11.546651, 14.599504, 3.562229, 8.775642, 4.254566, 6.718379, 8.507878, 16.240278, 5.473678, 12.803893, 5.945518, 13.063529, 4.806285, 10.476816, 5.405599, 11.772844, 8.246965, 14.829949, 10.433558, 8.690292, 20.060936, 11.21025, 1.815927, 1.501856, 27.158871, 52.405888, 55.166346, 4.749194, 7.490943, 7.530119, 2.022887, 4.246994, 5.590075, 2.691232, ... 4.114743, 12.67418, 7.105023, 2.177899, 26.163962, 34.280646, 10.047814, 3.597676, 14.217049, 15.178615, 3.358748, 3.103717, 3.402849, 14.731842, 43.983755, 44.554943, 3.439804, 22.717254, 25.98258, 23.190905, 34.407607, 26.470292, 17.409557, 11.20474, 62.043047, 58.04404, 40.144143, 3.193113, 4.539008, 4.354784, 1.036555, 6.741147, 4.326012, 3.721033, 7.992666, 13.336474, 2.023786, 10.900128, 18.255682, 10.512608, 4.763553, 10.223482, 12.434552, 7.13746, 7.779022, 14.35364, 5.405661, 1.799958, 3.141756, 8.433383, 64.531922, 63.856632, 2.449568, 6.878414, 9.087604, 4.639908, 4.746736, 0.842379, 1.267315, 3.331215, 1.941473, 5.580157, 2.335859, 8.794628, 5.67464, 34.289742, 5.754497, 4.672198, 3.74312, 0.779437, 2.702962, 5.760082, 6.195596, 6.631199, 0.004488, 0.621811, 4.285341, 0.913034, 0.38397, 35.787436, 41.785607, 0.961281, 11.162265, 17.989819, 8.353595, 15.668894, 35.943714, 22.63512, 11.558286, 3.220078, 58.012187, 24.28336, 6.679227, 43.867061, 5.32368, 0.495896, 6.311726, 24.498747, 25.813472, 37.393871, 38.187826, 26.260647, 15.95291, 63.187408, 0.072393, 2.570701, 2.136051, 18.593435, 4.1729, 0.012638, 0.00143, 0.778508, 6.036396, 19.793558, 37.282512, 8.192448, 48.146883, 70.024163, 0.527611], dtype=object) - mgrs:utm_zone()int6432
array(32)
- mgrs:latitude_band()<U1'T'
array('T', dtype='<U1') - s2:nodata_pixel_percentage(time)object0 11.344289 7e-05 ... 0 0 11.180457
array([0, 11.344289, 7e-05, 11.695041, 0, 11.747443, 0.001022, 11.865249, 0.002024, 11.836424, 0.000362, 10.520035, 11.043341, 4.3e-05, 10.245421, 12.794904, 0, 11.317464, 11.346283, 0, 11.001211, 0, 11.373752, 2.7e-05, 11.111417, 2e-05, 11.571965, 11.595662, 2.7e-05, 11.062833, 11.263861, 0, 11.629573, 0, 11.077637, 0, 11.491714, 0, 0.000849, 10.868026, 0, 11.355454, 0.000186, 0, 0, 11.066161, 11.089724, 0.000338, 0.007837, 10.673273, 10.849658, 2e-05, 0.000352, 10.598329, 10.622085, 0, 0.001427, 10.928832, 6.6e-05, 0.000524, 10.795923, 8e-05, 11.048497, 11.23339, 0, 0, 11.096473, 11.123971, 3e-06, 0, 11.194455, 11.381508, 7e-06, 0, 11.282003, 3e-06, 11.156502, 11.359793, 0.00151, 0, 11.419458, 0.000455, 11.023819, 0.001437, 11.483508, 11.501435, 0.000498, 2.3e-05, 10.923424, 11.122103, 0.00144, 0, 11.415191, 11.442321, 0.000488, 0.002694, 10.717951, 10.946921, 1.3e-05, 0, 11.634035, 11.675496, 0.000587, 0.008862, 41.191188, 53.534025, 41.394857, 53.682548, 4e-05, 3e-06, 11.886858, 0.000166, 0.00781, 10.717805, 10.919227, 2e-05, 12.038583, 12.075976, 0, 0, 10.400825, 2.7e-05, 12.22479, 5.3e-05, 0.005796, 10.700449, 10.900345, 0.000242, 0, 11.405729, 0, 11.032376, 0, 11.439073, 3e-06, 11.349372, 0, 11.473005, 11.499272, 0.000182, 11.398316, 0.000474, 11.407556, 0, ... 11.174101, 11.206333, 3e-06, 3e-06, 11.498877, 11.734018, 0, 11.156423, 11.126715, 0, 7e-06, 11.874377, 11.646823, 0, 3e-06, 10.985302, 11.013988, 0, 0, 11.783398, 11.556501, 8.3e-05, 1.3e-05, 10.792472, 10.827903, 0, 1.7e-05, 11.396532, 11.627065, 0.000146, 1.7e-05, 10.879045, 10.92066, 0, 7e-06, 11.413001, 11.643498, 0, 7e-06, 11.108098, 11.074386, 3e-06, 11.529401, 3e-06, 11.233792, 0, 0, 11.535111, 0, 11.251618, 11.219913, 0, 0, 11.794078, 11.559985, 0, 0, 11.204886, 3.6e-05, 11.340842, 2e-05, 11.207345, 1.7e-05, 11.143825, 1.3e-05, 11.229714, 11.26765, 3e-05, 10.9286, 0, 2e-05, 14.280696, 14.348489, 0.00062, 7.6e-05, 10.944625, 11.172747, 0.00077, 0.000232, 10.84151, 10.876025, 0, 0, 10.601236, 10.82366, 3e-06, 5.6e-05, 11.168964, 0, 1e-05, 11.48045, 11.250049, 3e-06, 0.000166, 11.488724, 11.445553, 0, 3e-06, 11.772808, 11.547185, 7e-06, 0.000292, 11.806361, 11.772018, 0.000196, 12.032601, 11.805047, 8.3e-05, 11.74587, 11.77223, 0, 0, 12.020504, 3e-06, 2.7e-05, 11.587144, 11.623067, 0, 0, 11.874768, 11.338881, 0, 0, 11.508559, 1.3e-05, 11.090159, 0.000448, 11.260354, 2.7e-05, 11.277342, 11.249379, 0, 3e-06, 11.501093, 0, 66.107917, 66.058993, 27.93659, 0, 10.693993, 10.921072, 4.3e-05, 0.000471, 12.545553, 0, 0, 11.074379, 0, 0, 11.180457], dtype=object) - s2:datatake_id(time)<U34'GS2A_20161105T101212_007169_N00...
array(['GS2A_20161105T101212_007169_N00.01', 'GS2A_20161108T102232_007212_N00.01', 'GS2A_20161115T101302_007312_N00.01', 'GS2A_20161118T102322_007355_N00.01', 'GS2A_20161125T101342_007455_N00.01', 'GS2A_20161128T102352_007498_N00.01', 'GS2A_20161205T101402_007598_N00.01', 'GS2A_20161208T102422_007641_N00.01', 'GS2A_20161215T101422_007741_N00.01', 'GS2A_20161218T102432_007784_N00.01', 'GS2A_20161225T101432_007884_N00.01', 'GS2A_20161228T102432_007927_N00.01', 'GS2A_20170716T102021_010787_N00.01', 'GS2B_20180124T101309_004624_N00.01', 'GS2B_20180308T102019_005239_N00.01', 'GS2B_20180328T102019_005525_N02.06', 'GS2A_20180330T101021_014462_N02.06', 'GS2A_20180402T102021_014505_N02.06', 'GS2A_20180402T102021_014505_N05.00', 'GS2B_20180404T101019_005625_N02.06', ... 'GS2A_20211202T102401_033667_N03.01', 'GS2B_20211204T101309_024787_N05.00', 'GS2B_20211204T101309_024787_N03.01', 'GS2B_20211207T102319_024830_N03.01', 'GS2A_20211209T101411_033767_N03.01', 'GS2A_20211212T102431_033810_N05.00', 'GS2A_20211212T102431_033810_N03.01', 'GS2A_20211212T102431_033810_N03.01', 'GS2B_20211214T101329_024930_N03.01', 'GS2B_20211217T102329_024973_N03.01', 'GS2B_20211217T102329_024973_N05.00', 'GS2A_20211219T101431_033910_N05.00', 'GS2A_20211219T101431_033910_N03.01', 'GS2A_20211222T102441_033953_N03.01', 'GS2B_20211224T101329_025073_N05.00', 'GS2B_20211224T101329_025073_N03.01', 'GS2B_20211227T102339_025116_N03.01', 'GS2A_20211229T101431_034053_N05.00', 'GS2A_20211229T101431_034053_N03.01', 'GS2A_20220101T102431_034096_N03.01'], dtype='<U34') - earthsearch:boa_offset_applied(time)boolFalse False False ... False False
array([False, False, False, False, False, False, False, False, False, False, False, False, False, False, False, False, False, False, True, False, False, False, False, False, False, False, False, True, False, False, True, False, False, False, False, False, False, False, True, False, False, False, False, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, False, True, False, False, False, True, False, True, False, True, False, True, False, True, False, True, False, False, False, True, False, True, False, False, False, False, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, False, True, True, False, True, False, False, True, False, True, False, False, True, False, True, False, False, False, False, True, False, True, False, True, False, False, False, False, False, False, False, False, False, True, False, False, False, False, False, False, True, False, False, False, False, False, False, True, False, False, False, False, False, False, False, False, False, False, False, False, False, False, False, False, True, False, False, False, False, True, False, False, False, False, False, ... False, False, True, True, True, False, True, False, True, False, True, False, False, True, False, False, True, False, True, False, True, False, True, False, False, True, True, False, False, True, False, True, False, True, False, True, False, True, False, False, True, True, False, True, False, True, False, False, True, True, False, False, True, True, False, False, True, True, False, False, True, True, False, True, False, False, False, False, False, True, False, False, False, True, False, True, False, True, False, True, False, False, False, False, False, False, False, False, False, False, True, False, False, True, False, False, True, True, False, False, True, True, False, False, True, True, False, False, True, True, False, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, False, True, False, False, False, True, True, False, False, True, False, False, True, True, False, False, False, True, False, False, False, False, False, False, False, True, False, True, False, False, False, True, False, False, False, False, True, True, False, False, True, False, False, True, False, False]) - view:sun_azimuth(time)float64168.8 171.5 168.9 ... 165.4 167.4
array([168.76730404, 171.5007273 , 168.92490676, 171.43109832, 168.65069955, 170.96072959, 167.99361455, 170.15238598, 167.0355798 , 169.09208589, 165.86605548, 167.87440027, 151.8234341 , 162.06246073, 161.48055258, 161.03048343, 157.56730763, 160.9519171 , 160.95112871, 157.33202866, 160.81618265, 157.10148033, 160.68316154, 156.78913455, 160.46385391, 156.45750616, 160.2253509 , 160.22455127, 156.02263791, 159.87653507, 159.87574753, 155.55588033, 159.49306276, 154.95926474, 158.96790448, 154.32314214, 158.40143575, 153.54420903, 153.54347793, 157.68294592, 152.74715303, 156.95027634, 151.83088055, 150.95184353, 150.95114938, 155.27317745, 155.27244139, 150.00823813, 150.00754544, 154.39096022, 154.39023005, 149.15290636, 149.15223072, 153.58734423, 153.58664329, 148.33422881, 148.33358015, 152.83638728, 147.65420999, 147.65356829, 152.22550532, 147.1117255 , 151.74945113, 151.74878145, 146.77173476, 146.77110182, 151.47727705, 151.47658522, 146.60408966, 146.60343708, 151.36678146, 151.36608569, 146.67333877, 146.67269084, 151.49062048, 146.91503921, 151.77344674, 151.77278843, 147.40016803, 147.39956026, ... 161.7283535 , 158.54479104, 158.5441226 , 162.91418092, 162.91484991, 159.86303814, 159.86238306, 164.11643944, 164.11709335, 161.08839998, 161.08777043, 165.21262305, 165.21324365, 162.31470033, 162.31409595, 166.30679222, 163.40604336, 163.40545498, 167.26091541, 167.26033544, 164.4675583 , 164.46699671, 168.18001689, 168.17946164, 165.3802784 , 165.37972912, 168.95169412, 168.95114043, 166.23954491, 166.23900125, 169.66922933, 169.66869182, 166.94828604, 170.24027851, 170.23974912, 167.58818255, 170.7407418 , 170.74126393, 168.07724029, 168.07672634, 171.09431652, 168.48147814, 168.4809826 , 171.36571904, 171.36621844, 168.73734455, 168.73684225, 171.49581873, 171.5431442 , 168.93545764, 168.93496557, 171.46404312, 168.88151331, 171.31062082, 168.70337048, 171.03549914, 168.46003736, 170.70845873, 170.70798852, 168.09340887, 168.09293917, 170.26759368, 167.67922239, 169.79854654, 169.79808633, 169.79808633, 167.16728228, 169.2357253 , 169.23619963, 166.6458662 , 166.64539797, 168.6820382 , 166.0305166 , 166.03005272, 168.04416312, 165.42923999, 165.42876808, 167.43292429]) - s2:datastrip_id(time)<U64'S2A_OPER_MSI_L2A_DS_SHIT_201905...
array(['S2A_OPER_MSI_L2A_DS_SHIT_20190504T200851_S20161105T101257_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190504T224516_S20161108T102425_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190504T225049_S20161115T101301_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190505T012451_S20161118T102318_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190505T004539_S20161125T101340_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190505T013016_S20161128T102354_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190505T201826_S20161205T101412_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190505T185759_S20161208T102418_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190505T054132_S20161215T101510_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190505T143721_S20161218T102606_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190505T122049_S20161225T101625_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20190508T201139_S20161228T102428_N00.01', 'S2A_OPER_MSI_L2A_DS_SHIT_20201008T134008_S20170716T102324_N00.01', 'S2B_OPER_MSI_L2A_DS_SHIT_20200930T170455_S20180124T101352_N00.01', 'S2B_OPER_MSI_L2A_DS_SHIT_20200315T002708_S20180308T102241_N00.01', 'S2B_OPER_MSI_L2A_DS_SGS__20180328T155221_S20180328T102738_N02.07', 'S2A_OPER_MSI_L2A_DS_EPAE_20180330T110651_S20180330T101620_N02.07', 'S2A_OPER_MSI_L2A_DS_SGS__20180402T155007_S20180402T102435_N02.07', 'S2A_OPER_MSI_L2A_DS_S2RP_20230908T074848_S20180402T102435_N05.00', 'S2B_OPER_MSI_L2A_DS_EPAE_20180404T113026_S20180404T101021_N02.07', ... 'S2B_OPER_MSI_L2A_DS_S2RP_20221223T132023_S20211204T101631_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20211204T130603_S20211204T101631_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211207T122554_S20211207T102344_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS4_20211209T115742_S20211209T101415_N03.01', 'S2A_OPER_MSI_L2A_DS_S2RP_20221224T052549_S20211212T102427_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20211212T115021_S20211212T102427_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211212T131717_S20211212T102745_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211214T120410_S20211214T101409_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211217T122256_S20211217T102332_N03.01', 'S2B_OPER_MSI_L2A_DS_S2RP_20221228T160151_S20211217T102332_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20221227T163048_S20211219T101431_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20211219T120654_S20211219T101431_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211222T131613_S20211222T102744_N03.01', 'S2B_OPER_MSI_L2A_DS_S2RP_20221228T143406_S20211224T101331_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20211224T123805_S20211224T101331_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211227T123108_S20211227T102359_N03.01', 'S2A_OPER_MSI_L2A_DS_S2RP_20221227T034720_S20211229T101430_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20211229T120455_S20211229T101430_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20220101T133200_S20220101T102607_N03.01'], dtype='<U64') - s2:unclassified_percentage(time)object0.000577 4.827457 ... 5.033683
array([0.000577, 4.827457, 5.253749, 0.925009, 0.085458, 5.489837, 4.052247, 4.710426, 4.071144, 5.195199, 6.043316, 4.808655, 1.717609, 5.420588, 9.280977, 8.551812, 2.35049, 7.303479, 0.303118, 0.210845, 7.539508, 0.046645, 0, 9.475724, 5.456717, 5.909364, 5.594448, 0.371043, 5.466987, 3.948615, 1.463659, 2.923919, 0.010591, 2.110809, 3.03697, 3.474819, 3.423198, 1.208596, 1e-05, 2.712885, 0.293834, 0.488316, 3.977711, 4.454073, 1.160262, 4.358478, 1.098276, 1.069784, 3.163287, 4.544741, 2.681325, 4.460995, 2.005034, 2.43801, 1.980482, 0.034416, 0, 3.439552, 3.927159, 0.997523, 4.376997, 2.976867, 4.058516, 1.663498, 0.541936, 0.006712, 4.843621, 1.244462, 5.249153, 2.221008, 1.996426, 0.862046, 4.255232, 1.372819, 6.023263, 4.26336, 5.166278, 3.875099, 3.965972, 0.934582, 0.528821, 4.920238, 4.501995, 3.978103, 4.030671, 1.056863, 7.394481, 4.603241, 4.559057, 1.179022, 3.255965, 1.28756, 6.709091, 2.610274, 5.480842, 2.305695, 6.044776, 1.711439, 5.925489, 1.566066, 5.030606, 2.001484, 6.699011, 3.156888, 5.373033, 4.150899, 2.856579, 2.486389, 3.317273, 0.482566, 4.919486, 1.418199, 0.617382, 5.468149, 2.296727, 6.54409, 4.218559, 0.61944, 4.174412, 0.527208, 8.282817, 7.037218, 4.932306, 5.010351, ... 0.770469, 0.292153, 0.427856, 3.074201, 3.535479, 0.935654, 0.376936, 2.392862, 3.128634, 1.085689, 0.59594, 3.233828, 3.988854, 4.071577, 3.205832, 3.502811, 3.476528, 0.901185, 1.40753, 0.903793, 4.482188, 6.716335, 1.300598, 2.973832, 1.438655, 2.57608, 4.269201, 6.485406, 0.020763, 1.156984, 0.000123, 3.403885, 4.25782, 4.223125, 2.17731, 6.710576, 4.584606, 4.550951, 6.723747, 3.530359, 3.061006, 2.525414, 2.565914, 5.964719, 4.865988, 1.982073, 1.313746, 3.965119, 7.643661, 3.9195, 0.708835, 2.723365, 3.860949, 1.847841, 0.0115, 0.311094, 4.260587, 0.578294, 2.210643, 5.30179, 3.644007, 0.124747, 2.356505, 0.831079, 4.109764, 0.574301, 4.626107, 2.220843, 3.195269, 1.787828, 3.676185, 2.024649, 5.294078, 0.072926, 4.682079, 0.765953, 4.788673, 4.617824, 0.000803, 6.356327, 4.773093, 7.133023, 0.034025, 0.018122, 0.836693, 8.035865, 2.179021, 3.463808, 6.254808, 0.659893, 0.012598, 0.874115, 5.403754, 5.800857, 4.6e-05, 0.018003, 7.35464, 3.775383, 0.822614, 5.32987, 2.792623, 4.462714, 0.132792, 0.485511, 2.107836, 4.74293, 6.752571, 0.001695, 0.007234, 5.996048, 6.082753, 0.215281, 4.698034, 0.001508, 0.001105, 4.838076, 5.000642, 0, 0.007372, 3.16737, 0.364003, 0.82729, 5.033683], dtype=object) - created(time)<U24'2022-11-08T11:45:48.722Z' ... '...
array(['2022-11-08T11:45:48.722Z', '2022-11-08T12:04:13.289Z', '2022-11-08T11:45:57.228Z', '2022-11-03T09:34:52.014Z', '2022-11-08T11:48:05.102Z', '2022-11-08T11:45:48.030Z', '2022-11-08T11:48:22.923Z', '2022-11-08T11:47:34.155Z', '2022-11-08T12:04:14.741Z', '2022-11-08T11:48:22.390Z', '2022-11-08T11:48:23.433Z', '2022-11-08T11:48:28.986Z', '2022-11-08T11:50:26.339Z', '2022-11-08T11:48:26.509Z', '2022-11-08T12:04:09.409Z', '2022-11-03T10:07:12.277Z', '2022-11-08T11:48:17.112Z', '2022-11-03T10:07:58.946Z', '2024-03-17T06:22:19.457Z', '2022-11-08T11:47:52.653Z', '2022-11-08T11:47:54.685Z', '2022-11-08T12:04:09.418Z', '2022-11-03T10:21:43.634Z', '2022-11-08T11:50:57.906Z', '2022-11-03T10:17:47.910Z', '2022-11-03T10:17:37.475Z', '2022-11-08T12:04:54.762Z', '2024-03-26T02:21:48.168Z', '2022-11-08T12:04:57.795Z', '2022-11-08T11:49:19.981Z', '2024-03-26T01:15:20.524Z', '2022-11-08T11:49:21.312Z', '2022-11-08T11:48:35.657Z', '2022-11-08T11:51:18.657Z', '2022-11-03T10:05:50.597Z', '2022-11-03T10:14:45.013Z', '2022-11-03T09:35:56.338Z', '2022-11-08T12:05:53.592Z', '2024-03-25T19:43:15.001Z', '2022-11-08T12:05:55.072Z', ... '2022-11-06T04:43:01.273Z', '2023-06-26T11:48:45.811Z', '2022-11-06T04:43:12.966Z', '2022-11-06T04:41:58.919Z', '2023-06-24T20:37:58.828Z', '2023-06-30T18:35:20.074Z', '2022-11-06T04:43:58.654Z', '2022-11-06T04:43:51.058Z', '2022-11-03T10:38:49.526Z', '2023-06-21T22:07:52.241Z', '2022-11-06T04:43:09.999Z', '2022-11-06T04:42:57.691Z', '2022-11-06T04:41:58.889Z', '2022-11-06T04:43:35.334Z', '2022-11-03T10:38:18.195Z', '2022-11-06T13:34:45.866Z', '2022-11-06T04:43:39.759Z', '2023-06-21T19:18:07.765Z', '2022-11-06T04:42:35.694Z', '2023-06-21T16:44:11.529Z', '2022-11-06T04:44:51.652Z', '2022-11-06T04:45:13.963Z', '2022-11-06T04:43:06.076Z', '2023-06-21T13:25:26.939Z', '2022-11-06T04:43:49.287Z', '2022-11-06T04:43:54.186Z', '2022-11-06T04:43:31.291Z', '2022-11-06T04:43:13.303Z', '2023-06-29T19:55:36.667Z', '2023-06-30T07:19:57.427Z', '2022-11-03T10:37:43.273Z', '2022-11-06T04:42:53.943Z', '2023-06-29T19:52:20.154Z', '2022-11-06T04:44:49.049Z', '2022-11-06T13:41:28.061Z', '2023-06-30T09:10:08.167Z', '2022-11-03T10:01:41.689Z', '2022-11-03T10:20:25.492Z'], dtype='<U24') - view:sun_elevation(time)float6426.99 26.4 24.21 ... 19.04 19.6
array([26.98508106, 26.39590817, 24.205512 , 23.75564628, 21.92260219, 21.65441999, 20.23442729, 20.18551916, 19.22450817, 19.42218358, 18.94706454, 19.40650105, 62.54811902, 22.5064948 , 37.09649697, 45.09552022, 45.28152111, 47.07100484, 47.07084319, 47.23197698, 49.00103354, 49.14342751, 50.88352597, 50.98806745, 52.69318301, 52.7654197 , 54.42804858, 54.42790188, 54.44826435, 56.06290503, 56.06276394, 56.03719768, 57.597432 , 57.50421503, 59.00614766, 58.85390576, 60.29208284, 60.05704583, 60.05688863, 61.42874688, 61.12319465, 62.42385603, 62.02300276, 62.77256339, 62.77241485, 63.92369553, 63.92357078, 63.34451662, 63.34436721, 64.41924796, 64.41912306, 63.75767591, 63.75752822, 64.75096851, 64.75084622, 63.99574815, 63.99560386, 64.90966251, 64.07279512, 64.07265122, 64.90532109, 63.98612537, 64.73636857, 64.73624903, 63.74897137, 63.74882853, 64.41612049, 64.41599831, 63.35876078, 63.35861527, 63.94197648, 63.94185525, 62.83388744, 62.83374528, 63.33223335, 62.16862801, 62.58230373, 62.58218939, 61.38682708, 61.38669456, 61.71510093, 60.47914107, 60.72217298, 59.47145257, 59.62764779, 59.62754935, 58.34875584, 58.34864006, 58.41870318, 58.41861182, 57.13847067, 57.1383626 , 57.12146807, 57.12138344, 55.82341318, 55.82331255, 55.72254359, 55.72246811, 54.4356281 , 54.43553891, ... 59.79652779, 58.52795949, 58.52775363, 58.60893721, 57.32281075, 57.31793899, 56.02938815, 55.9397754 , 54.64405186, 54.47195716, 53.1857231 , 52.93311116, 52.93325728, 51.64576322, 51.31583936, 50.04649385, 50.04634339, 49.64226531, 49.64239446, 48.37714277, 48.37700216, 47.90599319, 47.90611534, 46.66460356, 46.66446865, 46.13173754, 46.13185429, 44.90173756, 44.90160997, 44.31518713, 44.31529615, 43.11607325, 43.11595334, 42.481491 , 41.29749094, 41.29737536, 40.62479888, 40.62470019, 39.47363892, 39.47353057, 38.7697373 , 38.76964514, 37.63964619, 37.63954293, 36.916899 , 36.91681058, 35.82282842, 35.82272969, 35.09066166, 35.09057829, 34.02427289, 33.29583895, 33.29575963, 32.26912819, 31.55366216, 31.55373753, 30.56006843, 30.5599828 , 29.87006622, 28.91903429, 28.91895383, 28.26522552, 28.26529335, 27.35234395, 27.35226484, 26.7480921 , 25.33890244, 24.51698675, 24.51691171, 24.04735644, 23.27661249, 22.89071122, 22.16763464, 21.87538032, 21.20891488, 21.01932412, 21.01926647, 20.40271612, 20.40264773, 20.32515116, 19.76865853, 19.81067577, 19.81062012, 19.81062012, 19.30635163, 19.47330287, 19.47335998, 19.03402517, 19.03395836, 19.32933111, 18.93992201, 18.93985622, 19.36693127, 19.04271818, 19.04265123, 19.6012287 ]) - s2:water_percentage(time)object0 1.504413 ... 0.070587 5.664684
array([0, 1.504413, 1.718854, 0.077238, 0, 3.750497, 5.19834, 4.202895, 5.982503, 5.592868, 0.682943, 31.421277, 0.457302, 3.741326, 0.329686, 0.137873, 0.053789, 0.251412, 0.207097, 0.000843, 0.311848, 0.000474, 0, 0.187913, 0.748155, 0.268118, 0.917805, 0.346678, 1.027319, 0.116639, 0.073531, 0.005103, 0, 0.006377, 0.584103, 0.06419, 0.087478, 0.000388, 0.00065, 0.051894, 3e-05, 0, 0.085863, 0.040863, 0.071343, 0.182953, 0.225964, 4.6e-05, 0.002117, 0.071563, 0.106249, 0.08353, 0.131944, 0.005107, 0.014032, 0, 0, 0.235848, 0.161967, 0.163686, 0.30532, 0.296117, 0.199112, 0.302033, 0, 0.004386, 0.207728, 0.298044, 0.153642, 0.188865, 0.069618, 0.101877, 0.262637, 0.284372, 0.213024, 0.403837, 0.168186, 0.152903, 0.361408, 0.311333, 4.1e-05, 0.117853, 0.208107, 0.228894, 0.733601, 0.481042, 0.15578, 0.159923, 0.870363, 0.505945, 0.259873, 0.227644, 0.263255, 0.195666, 0.202605, 0.176418, 0.498155, 0.287794, 0.78802, 0.484242, 0.739641, 0.433322, 0.387519, 0.273139, 0.133371, 0.072996, 0.082435, 0.058574, 0.320885, 0.339086, 0.192214, 0.00496, 0.009858, 0.216817, 0.239804, 0.502102, 0.512012, 0.38304, 0.15999, 0.179624, 0.149878, 0.360799, 0.502129, 0.528028, 0.398496, 0.581379, 0.47856, 0.38205, 0.335327, 0.360114, 0, ... 0.405969, 0.275791, 0.281174, 0.337965, 0.191665, 0.120455, 0.274382, 0.366963, 0.120382, 0.116535, 0.106947, 0.460107, 0.255931, 0.011337, 0.013162, 0.013852, 0.417062, 0.407079, 0.083903, 0.093668, 0.016616, 0.02353, 0.187265, 0.291207, 0.001825, 0.008733, 1.5e-05, 0.426784, 0.530594, 0.325099, 0.656142, 0.434982, 0.433164, 0.55737, 0.285478, 0.198213, 0.470105, 0.022566, 0.22063, 0.346635, 0.42016, 0.283609, 0.299676, 0.434694, 0.421726, 0.27406, 0.343197, 0.464824, 0.592808, 0.392457, 0.004436, 0.003049, 0.497876, 0.364553, 0.23904, 0.351266, 0.267654, 0.222501, 0.239024, 0.227711, 0.290617, 0.283393, 0.36281, 0.034044, 0.050381, 0.065322, 0.112598, 0.265469, 0.392738, 0.324478, 0.42346, 0.337052, 0.474906, 0.347677, 0.361514, 0.549882, 0.309956, 0.617706, 0.348512, 0.003587, 0, 0.796821, 0.115922, 0.150799, 0.66719, 0.222747, 0.007538, 0.025355, 1.622207, 1.623247, 0.002249, 0.002329, 1.334601, 0.18057, 0.005053, 3.737868, 0.863894, 0.793574, 0.030249, 0.071284, 0.061961, 0.386873, 2.633459, 0, 0.097845, 5.355824, 5.623126, 1.009572, 5.89586, 0.336842, 0.317886, 6.49615, 6.159603, 0.00153, 3e-06, 2.908082, 0.009015, 0.070587, 5.664684], dtype=object) - s2:sequence(time)<U1'0' '0' '0' '0' ... '0' '1' '0' '0'
array(['0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '1', '0', '0', '0', '0', '0', '0', '0', '0', '1', '0', '0', '1', '0', '0', '0', '0', '0', '0', '0', '1', '0', '0', '0', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '0', '1', '0', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '3', '0', '0', '0', '1', '0', '1', '0', '0', '0', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '1', '0', '3', '2', '0', '1', '0', '0', '1', '0', '1', '0', '0', '1', '0', '1', '0', '0', '0', '0', '1', '0', '1', '0', '1', '0', '0', '0', '0', '0', '0', '0', '0', '0', '1', '0', '0', '1', '0', '0', '0', '1', '0', '0', '0', '1', '0', '0', '1', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '1', '0', '0', '0', '0', '1', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '1', '0', '1', '0', '0', '1', '0', '0', '0', '0', '0', '1', '0', '1', '0', '3', '1', '1', '0', '1', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '0', '1', '0', '1', '0', '1', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '0', ... '1', '3', '1', '0', '1', '0', '0', '1', '1', '0', '0', '0', '1', '1', '0', '1', '0', '0', '1', '0', '3', '1', '0', '0', '1', '0', '1', '0', '0', '1', '0', '1', '1', '2', '0', '3', '0', '1', '0', '1', '0', '1', '0', '0', '3', '1', '0', '0', '1', '1', '0', '0', '1', '0', '0', '0', '1', '2', '0', '0', '1', '1', '0', '1', '0', '0', '0', '2', '0', '1', '0', '2', '0', '3', '0', '4', '1', '0', '0', '1', '2', '1', '0', '1', '0', '2', '0', '1', '0', '0', '1', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0', '1', '0', '0', '0', '0', '0', '1', '0', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '1', '0', '0', '1', '0', '1', '4', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0', '1', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '2', '1', '0', '1', '0', '0', '0', '3', '1', '0', '0', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0', '0', '0', '0', '0', '0', '0', '1', '0', '1', '0', '0', '0', '2', '1', '0', '0', '0', '1', '1', '0', '0', '1', '0', '0', '1', '0', '0'], dtype='<U1') - s2:high_proba_clouds_percentage(time)object24.661314 22.209878 ... 1.115038
array([24.661314, 22.209878, 6.867649, 79.345798, 94.944388, 3.168646, 0.745553, 0.413493, 1.174874, 1.170849, 24.156788, 1.374512, 1.337166, 0.367491, 0.570044, 26.703706, 78.384817, 1.773553, 1.234293, 87.978148, 13.401236, 76.867789, 33.670625, 1.490344, 26.342592, 0.518996, 2.820556, 2.205384, 16.116267, 68.850702, 68.15961, 67.601901, 36.8002, 73.803949, 47.75894, 66.505671, 71.743435, 84.642434, 64.6855, 70.836842, 65.79107, 50.946313, 60.565233, 69.637221, 69.338697, 42.098665, 41.678733, 31.702426, 31.403744, 52.695704, 52.652884, 45.989439, 45.187357, 40.025982, 39.917395, 3.208765, 3.052605, 8.267286, 40.882567, 40.489361, 26.83138, 3.955723, 13.401571, 12.273046, 48.849538, 48.705837, 19.129595, 18.43373, 31.176442, 30.647099, 16.919534, 16.826519, 12.891272, 11.998865, 43.324599, 10.209618, 22.121769, 22.033468, 10.861061, 10.389487, 81.155246, 50.657958, 34.052539, 30.774882, 10.181027, 9.899831, 44.513047, 44.860804, 14.938051, 14.835966, 48.084, 47.945249, 50.272238, 50.233287, 55.757356, 55.903018, 31.791651, 31.738433, 14.597157, 14.290924, 25.078273, 24.989153, 37.233722, 37.379235, 62.81786, 77.758723, 62.756652, 78.034508, 1.557822, 1.011191, 52.984357, 40.419045, 39.954108, 16.039495, 15.155551, 34.752625, 4.432537, 4.220365, 0.403691, 0.125501, ... 9.307959, 39.434332, 39.099041, 20.228285, 20.820685, 47.36017, 47.621009, 65.818799, 18.608476, 6.247902, 79.256183, 47.700036, 48.553547, 9.08678, 36.520809, 60.108793, 60.104287, 54.052699, 54.310071, 31.285718, 30.789036, 35.123378, 35.160905, 53.882593, 14.315453, 15.461715, 22.547159, 1.890319, 33.023441, 24.895471, 11.225515, 43.364966, 43.255213, 5.183531, 75.152022, 27.849272, 28.16529, 16.227871, 15.446132, 19.057457, 19.14739, 38.547277, 38.847998, 7.280029, 7.486678, 14.403863, 14.277443, 34.561864, 35.447484, 9.287885, 9.255257, 18.762164, 19.340248, 8.849865, 0.345805, 0.561631, 3.4368, 3.498174, 7.280371, 7.562798, 84.947991, 84.994805, 49.405092, 49.980921, 3.932654, 5.505205, 0.654732, 1.49993, 7.433846, 8.423661, 3.173025, 0.003402, 0.556202, 0.676435, 0.767157, 0.338008, 55.403703, 55.54024, 0.398867, 24.077275, 24.494292, 14.072886, 13.744904, 59.666604, 60.881168, 0.41675, 1.170431, 37.360683, 38.054952, 24.5204, 6.742643, 71.681809, 1.315025, 53.176111, 11.245214, 41.957614, 42.616093, 11.846358, 12.814035, 7.818661, 35.913226, 0.003612, 1.365308, 1.818938, 65.659213, 1.222941, 0.018962, 0.001032, 1.436024, 1.420145, 57.708973, 59.827262, 39.617011, 19.537075, 20.354392, 1.115038], dtype=object) - title(band)<U31'Aerosol optical thickness (AOT)...
array(['Aerosol optical thickness (AOT)', 'Blue (band 2) - 10m', 'Coastal aerosol (band 1) - 60m', 'Green (band 3) - 10m', 'NIR 1 (band 8) - 10m', 'NIR 2 (band 8A) - 20m', 'NIR 3 (band 9) - 60m', 'Red (band 4) - 10m', 'Red edge 1 (band 5) - 20m', 'Red edge 2 (band 6) - 20m', 'Red edge 3 (band 7) - 20m', 'Scene classification map (SCL)', 'SWIR 1 (band 11) - 20m', 'SWIR 2 (band 12) - 20m', 'True color image', 'Water vapour (WVP)', 'Aerosol optical thickness (AOT)', 'Blue (band 2) - 10m', 'Coastal aerosol (band 1) - 60m', 'Green (band 3) - 10m', 'NIR 1 (band 8) - 10m', 'NIR 2 (band 8A) - 20m', 'NIR 3 (band 9) - 60m', 'Red (band 4) - 10m', 'Red edge 1 (band 5) - 20m', 'Red edge 2 (band 6) - 20m', 'Red edge 3 (band 7) - 20m', 'Scene classification map (SCL)', 'SWIR 1 (band 11) - 20m', 'SWIR 2 (band 12) - 20m', 'True color image', 'Water vapour (WVP)'], dtype='<U31') - raster:bands(band)object[{'nodata': 0, 'data_type': 'uin...
array([list([{'nodata': 0, 'data_type': 'uint16', 'bits_per_sample': 15, 'spatial_resolution': 20, 'scale': 0.001, 'offset': 0}]), None, None, None, None, None, None, None, None, None, None, list([{'nodata': 0, 'data_type': 'uint8', 'spatial_resolution': 20}]), None, None, None, list([{'nodata': 0, 'data_type': 'uint16', 'bits_per_sample': 15, 'spatial_resolution': 20, 'unit': 'cm', 'scale': 0.001, 'offset': 0}]), list([{'nodata': 0, 'data_type': 'uint16', 'bits_per_sample': 15, 'spatial_resolution': 20, 'scale': 0.001, 'offset': 0}]), None, None, None, None, None, None, None, None, None, None, list([{'nodata': 0, 'data_type': 'uint8', 'spatial_resolution': 20}]), None, None, None, list([{'nodata': 0, 'data_type': 'uint16', 'bits_per_sample': 15, 'spatial_resolution': 20, 'unit': 'cm', 'scale': 0.001, 'offset': 0}])], dtype=object) - gsd(band)objectNone 10 60 10 ... 20 20 None None
array([None, 10, 60, 10, 10, 20, 60, 10, 20, 20, 20, None, 20, 20, None, None, None, 10, 60, 10, 10, 20, 60, 10, 20, 20, 20, None, 20, 20, None, None], dtype=object) - common_name(band)objectNone 'blue' 'coastal' ... None None
array([None, 'blue', 'coastal', 'green', 'nir', 'nir08', 'nir09', 'red', 'rededge', 'rededge', 'rededge', None, 'swir16', 'swir22', None, None, None, 'blue', 'coastal', 'green', 'nir', 'nir08', 'nir09', 'red', 'rededge', 'rededge', 'rededge', None, 'swir16', 'swir22', None, None], dtype=object) - center_wavelength(band)objectNone 0.49 0.443 ... 2.19 None None
array([None, 0.49, 0.443, 0.56, 0.842, 0.865, 0.945, 0.665, 0.704, 0.74, 0.783, None, 1.61, 2.19, None, None, None, 0.49, 0.443, 0.56, 0.842, 0.865, 0.945, 0.665, 0.704, 0.74, 0.783, None, 1.61, 2.19, None, None], dtype=object) - full_width_half_max(band)objectNone 0.098 0.027 ... None None
array([None, 0.098, 0.027, 0.045, 0.145, 0.033, 0.026, 0.038, 0.019, 0.018, 0.028, None, 0.143, 0.242, None, None, None, 0.098, 0.027, 0.045, 0.145, 0.033, 0.026, 0.038, 0.019, 0.018, 0.028, None, 0.143, 0.242, None, None], dtype=object) - epsg()int6432632
array(32632)
- timePandasIndex
PandasIndex(DatetimeIndex(['2016-11-05 10:12:57.363000', '2016-11-08 10:24:25.911000', '2016-11-15 10:13:01.462000', '2016-11-18 10:23:18.464000', '2016-11-25 10:13:40.461000', '2016-11-28 10:23:54.458000', '2016-12-05 10:14:12.679000', '2016-12-08 10:24:18.464000', '2016-12-15 10:15:10.357000', '2016-12-18 10:26:06.867000', ... '2021-12-17 10:27:49.271000', '2021-12-19 10:17:59.960000', '2021-12-19 10:17:59.962000', '2021-12-22 10:27:56.749000', '2021-12-24 10:17:54.425000', '2021-12-24 10:17:54.425000', '2021-12-27 10:27:51.006000', '2021-12-29 10:18:00.908000', '2021-12-29 10:18:00.910000', '2022-01-01 10:27:57.174000'], dtype='datetime64[ns]', name='time', length=966, freq=None)) - bandPandasIndex
PandasIndex(Index(['aot', 'blue', 'coastal', 'green', 'nir', 'nir08', 'nir09', 'red', 'rededge1', 'rededge2', 'rededge3', 'scl', 'swir16', 'swir22', 'visual', 'wvp', 'aot-jp2', 'blue-jp2', 'coastal-jp2', 'green-jp2', 'nir-jp2', 'nir08-jp2', 'nir09-jp2', 'red-jp2', 'rededge1-jp2', 'rededge2-jp2', 'rededge3-jp2', 'scl-jp2', 'swir16-jp2', 'swir22-jp2', 'visual-jp2', 'wvp-jp2'], dtype='object', name='band')) - xPandasIndex
PandasIndex(Index([661130.0, 661140.0, 661150.0, 661160.0, 661170.0, 661180.0, 661190.0, 661200.0, 661210.0, 661220.0, ... 693130.0, 693140.0, 693150.0, 693160.0, 693170.0, 693180.0, 693190.0, 693200.0, 693210.0, 693220.0], dtype='float64', name='x', length=3210)) - yPandasIndex
PandasIndex(Index([5152650.0, 5152640.0, 5152630.0, 5152620.0, 5152610.0, 5152600.0, 5152590.0, 5152580.0, 5152570.0, 5152560.0, ... 5107400.0, 5107390.0, 5107380.0, 5107370.0, 5107360.0, 5107350.0, 5107340.0, 5107330.0, 5107320.0, 5107310.0], dtype='float64', name='y', length=4535))
- spec :
- RasterSpec(epsg=32632, bounds=(661130, 5107300, 693230, 5152650), resolutions_xy=(10, 10))
- crs :
- epsg:32632
- transform :
- | 10.00, 0.00, 661130.00| | 0.00,-10.00, 5152650.00| | 0.00, 0.00, 1.00|
- resolution :
- 10
From the output of the previous cell you can notice something really interesting: the size of the selected data is more than 3 TB!
But you should have noticed that it was too quick to download this huge amount of data.
This is what lazy loading allows: getting all the information about the data in a quick manner without having to access and download all the available files.
Quiz hint: look carefully at the dimensions of the loaded datacube!
Data Formats and Performance#
Chunking Explained#
When working with large Earth Observation datasets, it’s usually not possible to load all data into a computer’s memory at once. Instead, data is divided into smaller, manageable chunks. Each chunk is the smallest unit that can be processed independently. This chunking strategy enables efficient, piecewise data access and parallel processing, improving performance and scalability.
The figure below visually explains the concept of data chunking: on the left, a three-dimensional data cube (x, y, and time) is shown without chunks, while on the right, the same data cube is displayed with chunks highlighted.
Data Cube without chunking |
Data Cube with chunking |
|---|---|
|
|
Figure: on the left, a 3D data cube without any chunking strategy applied. On the right, a 3D data cube with box chunking.
There are several ways to chunk data depending on the analysis needs:
Spatial chunking divides data by geographic regions (e.g., longitude and latitude), ideal for spatial queries.
Time-based chunking divides datasets along temporal dimensions, useful for time series analysis.
Box chunking partitions data into fixed-size, 3D blocks combining space and time.
Choosing an appropriate chunking strategy is crucial, as it impacts read performance, memory usage, and cost-effectiveness in cloud environments.
Q&A: What Did You Discover?#
3 minutes
Key Concepts Participants Should Understand:
Memory Efficiency: Chunking enables processing datasets larger than RAM
Lazy Evaluation: Build complex operations without immediate computation
Cloud-Native: Stream only needed data chunks from remote storage
Scalability: Same patterns work from laptop to supercomputer
Questions for Understanding:
“How does chunking solve the ‘big data’ problem?”
“Why is lazy evaluation important for satellite data analysis?”