Some tips with xarray and pandas
Contents
Some tips with xarray and pandas#
We have massively different levels here
Try to make some aims for technical skills you can learn!
If you are beginning with python –> learn the basics
If you are good at basic python –> learn new packages
If you know all the packages –> improve your skills with producing your own software etc.
If you don’t know git and github –> get better at this!
print(‘Hey world’)#
Please feel free to come with suggestions and extra input as we go! Questions:How many have used jupyter lab?
How many have used pandas?
How many have used xarray?
What are pandas and xarray?#
Pandas –> like a spreadsheet 2D data with columns and rows
xarray –> like pandas, but in N dimensions
Use the functionality these packages gives you! Will help you avoid mistakes. Try to get as good as possible :)
Pandas#
 (Source: https://www.geeksforgeeks.org/python-pandas-dataframe/)
(Source: https://www.geeksforgeeks.org/python-pandas-dataframe/)
Xarray#
 (Source: https://docs.xarray.dev/)
(Source: https://docs.xarray.dev/)
1. Read in CMIP6 data: We will skip this next part, but you can check it later to read data:#
Import python packages#
import xarray as xr
xr.set_options(display_style='html')
import intake
import cftime
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import numpy as np
import pandas as pd
import datetime
import seaborn as sns
Open CMIP6 online catalog#
cat_url = "https://storage.googleapis.com/cmip6/pangeo-cmip6.json"
col = intake.open_esm_datastore(cat_url)
col
pangeo-cmip6 catalog with 7674 dataset(s) from 514818 asset(s):
| unique | |
|---|---|
| activity_id | 18 |
| institution_id | 36 |
| source_id | 88 |
| experiment_id | 170 |
| member_id | 657 |
| table_id | 37 |
| variable_id | 700 |
| grid_label | 10 |
| zstore | 514818 |
| dcpp_init_year | 60 |
| version | 736 |
Search corresponding data#
Please check here for info about CMIP and variables :)
Particularly useful is maybe the variable search which you find here: https://clipc-services.ceda.ac.uk/dreq/mipVars.html
cat = col.search(source_id=['CESM2'], experiment_id=['historical'], table_id=['Amon','fx','AERmon'],
variable_id=['tas','hurs', 'areacella','mmrso4' ], member_id=['r1i1p1f1'])
cat.df
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | zstore | dcpp_init_year | version | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CMIP | NCAR | CESM2 | historical | r1i1p1f1 | fx | areacella | gn | gs://cmip6/CMIP6/CMIP/NCAR/CESM2/historical/r1... | NaN | 20190308 |
| 1 | CMIP | NCAR | CESM2 | historical | r1i1p1f1 | AERmon | mmrso4 | gn | gs://cmip6/CMIP6/CMIP/NCAR/CESM2/historical/r1... | NaN | 20190308 |
| 2 | CMIP | NCAR | CESM2 | historical | r1i1p1f1 | Amon | hurs | gn | gs://cmip6/CMIP6/CMIP/NCAR/CESM2/historical/r1... | NaN | 20190308 |
| 3 | CMIP | NCAR | CESM2 | historical | r1i1p1f1 | Amon | tas | gn | gs://cmip6/CMIP6/CMIP/NCAR/CESM2/historical/r1... | NaN | 20190308 |
Create dictionary from the list of datasets we found#
This step may take several minutes so be patient!
dset_dict = cat.to_dataset_dict(zarr_kwargs={'use_cftime':True})
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
list(dset_dict.keys())
['CMIP.NCAR.CESM2.historical.fx.gn',
'CMIP.NCAR.CESM2.historical.Amon.gn',
'CMIP.NCAR.CESM2.historical.AERmon.gn']
Open dataset#
Use
xarraypython package to analyze netCDF datasetopen_datasetallows to get all the metadata without loading data into memory.with
xarray, we only load into memory what is needed.
ds1 = dset_dict['CMIP.NCAR.CESM2.historical.Amon.gn']
ds2 = dset_dict['CMIP.NCAR.CESM2.historical.fx.gn']
ds3 = dset_dict['CMIP.NCAR.CESM2.historical.AERmon.gn']
ds1
<xarray.Dataset>
Dimensions: (member_id: 1, time: 1980, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
lat_bnds (lat, nbnd) float32 dask.array<chunksize=(192, 2), meta=np.ndarray>
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float32 dask.array<chunksize=(288, 2), meta=np.ndarray>
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1980, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
Dimensions without coordinates: nbnd
Data variables:
hurs (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 600, 192, 288), meta=np.ndarray>
tas (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 600, 192, 288), meta=np.ndarray>
Attributes: (12/47)
physics_index: 1
sub_experiment_id: none
realization_index: 1
parent_experiment_id: piControl
branch_time_in_parent: 219000.0
cesm_casename: b.e21.BHIST.f09_g17.CMIP6-historical.001
... ...
parent_variant_label: r1i1p1f1
forcing_index: 1
parent_mip_era: CMIP6
external_variables: areacella
data_specs_version: 01.00.29
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.Amon.gnds3
<xarray.Dataset>
Dimensions: (lev: 32, nbnd: 2, lat: 192, lon: 288, member_id: 1, time: 1980)
Coordinates:
a_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
b_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
lat_bnds (lat, nbnd) float64 dask.array<chunksize=(192, 2), meta=np.ndarray>
* lev (lev) float64 -3.643 -7.595 -14.36 ... -957.5 -976.3 -992.6
lev_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1980, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
Dimensions without coordinates: nbnd
Data variables:
a (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
b (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
mmrso4 (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 10, 32, 192, 288), meta=np.ndarray>
p0 float32 ...
ps (time, lat, lon) float32 dask.array<chunksize=(10, 192, 288), meta=np.ndarray>
Attributes: (12/50)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-11-04;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/068116b9-bea7-4f4d-a942-e0bbee279412
version_id: v20190308
intake_esm_varname: ['mmrso4']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.AERmon.gnds_list = [dset_dict[l].drop('lat_bnds') for l in dset_dict.keys()]
ds = xr.merge(ds_list)
ds
<xarray.Dataset>
Dimensions: (member_id: 1, lat: 192, lon: 288, nbnd: 2, time: 1980, lev: 32)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1980, 2), meta=np.ndarray>
a_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
b_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* lev (lev) float64 -3.643 -7.595 -14.36 ... -957.5 -976.3 -992.6
lev_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
areacella (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
hurs (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 600, 192, 288), meta=np.ndarray>
tas (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 600, 192, 288), meta=np.ndarray>
a (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
b (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
mmrso4 (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 10, 32, 192, 288), meta=np.ndarray>
p0 float32 ...
ps (time, lat, lon) float32 dask.array<chunksize=(10, 192, 288), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gn1.1 Reading in the data from file:#
path='filename.nc'
ds = xr.open_dataset(path)
Opening multiple files:#
list_of_files = [
'file1.nc',
'file2.nc'
]
xr.open_mfdataset(list_of_files, concat_dim='time',combine='by_coords')
2. Check how your dataset looks#
Different types of information/data:#
Coordinates
Data variables
Global attributes
Variable attributes
ds
<xarray.Dataset>
Dimensions: (member_id: 1, lat: 192, lon: 288, nbnd: 2, time: 1980, lev: 32)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1980, 2), meta=np.ndarray>
a_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
b_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* lev (lev) float64 -3.643 -7.595 -14.36 ... -957.5 -976.3 -992.6
lev_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
areacella (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
hurs (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 600, 192, 288), meta=np.ndarray>
tas (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 600, 192, 288), meta=np.ndarray>
a (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
b (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
mmrso4 (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 10, 32, 192, 288), meta=np.ndarray>
p0 float32 ...
ps (time, lat, lon) float32 dask.array<chunksize=(10, 192, 288), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gn3. Sometimes we want to do some nice tweaks before we start:#
Selecting data and super quick plotting:#
xarray loads data only when it needs to (it’s lazy, Anne can explain), and you might want to early on define the subset of data you want to look at so that you don’t end up loading a lot of extra data.
See here for nice overview#
In order to reduce unecessary calculations and loading of data, think about which part of the data you want, and slice early on.
Slice in time: the sel method#
ds
<xarray.Dataset>
Dimensions: (member_id: 1, lat: 192, lon: 288, nbnd: 2, time: 1980, lev: 32)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1980, 2), meta=np.ndarray>
a_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
b_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* lev (lev) float64 -3.643 -7.595 -14.36 ... -957.5 -976.3 -992.6
lev_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
areacella (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
hurs (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 600, 192, 288), meta=np.ndarray>
tas (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 600, 192, 288), meta=np.ndarray>
a (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
b (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
mmrso4 (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 10, 32, 192, 288), meta=np.ndarray>
p0 float32 ...
ps (time, lat, lon) float32 dask.array<chunksize=(10, 192, 288), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gndss = ds.sel(time = slice('1990-01-01','2010-01-01'))
dss
<xarray.Dataset>
Dimensions: (member_id: 1, lat: 192, lon: 288, nbnd: 2, time: 240, lev: 32)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) object 1990-01-15 12:00:00 ... 2009-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(240, 2), meta=np.ndarray>
a_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
b_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* lev (lev) float64 -3.643 -7.595 -14.36 ... -957.5 -976.3 -992.6
lev_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
areacella (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
hurs (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 120, 192, 288), meta=np.ndarray>
tas (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 120, 192, 288), meta=np.ndarray>
a (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
b (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
mmrso4 (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 10, 32, 192, 288), meta=np.ndarray>
p0 float32 ...
ps (time, lat, lon) float32 dask.array<chunksize=(10, 192, 288), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gnYou might even select only the arctic e.g.:
isel, sel: index selecting#
Select the surface (which in this case is the last index :)
dss
<xarray.Dataset>
Dimensions: (member_id: 1, lat: 192, lon: 288, nbnd: 2, time: 240, lev: 32)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) object 1990-01-15 12:00:00 ... 2009-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(240, 2), meta=np.ndarray>
a_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
b_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* lev (lev) float64 -3.643 -7.595 -14.36 ... -957.5 -976.3 -992.6
lev_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
areacella (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
hurs (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 120, 192, 288), meta=np.ndarray>
tas (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 120, 192, 288), meta=np.ndarray>
a (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
b (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
mmrso4 (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 10, 32, 192, 288), meta=np.ndarray>
p0 float32 ...
ps (time, lat, lon) float32 dask.array<chunksize=(10, 192, 288), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gndss_s = dss.isel(lev=-1)
dss_s
<xarray.Dataset>
Dimensions: (member_id: 1, lat: 192, lon: 288, nbnd: 2, time: 240)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) object 1990-01-15 12:00:00 ... 2009-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(240, 2), meta=np.ndarray>
a_bnds (nbnd) float64 dask.array<chunksize=(2,), meta=np.ndarray>
b_bnds (nbnd) float64 dask.array<chunksize=(2,), meta=np.ndarray>
lev float64 -992.6
lev_bnds (nbnd) float64 dask.array<chunksize=(2,), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
areacella (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
hurs (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 120, 192, 288), meta=np.ndarray>
tas (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 120, 192, 288), meta=np.ndarray>
a float64 dask.array<chunksize=(), meta=np.ndarray>
b float64 dask.array<chunksize=(), meta=np.ndarray>
mmrso4 (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 10, 192, 288), meta=np.ndarray>
p0 float32 ...
ps (time, lat, lon) float32 dask.array<chunksize=(10, 192, 288), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
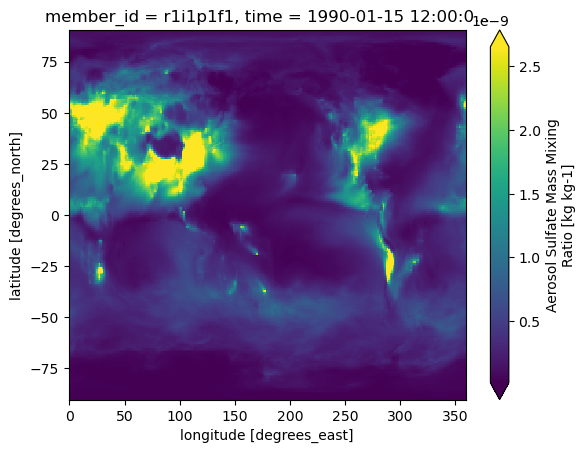
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gndss_s['mmrso4'].isel(time=0).plot(robust=True)
<matplotlib.collections.QuadMesh at 0x7f9573e49310>
3.2 Calculates variables and assign attributes!#
Nice for plotting and to keep track of what is in your dataset (especially ‘units’ and ‘standard_name’/’long_name’ will be looked for by xarray.
dss['T_C'] = dss['tas']-273.15
dss['T_C'] = dss['T_C'].assign_attrs({'units': '$^\circ$C'})
dss['T_C']
<xarray.DataArray 'T_C' (member_id: 1, time: 240, lat: 192, lon: 288)>
dask.array<sub, shape=(1, 240, 192, 288), dtype=float32, chunksize=(1, 120, 192, 288), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) object 1990-01-15 12:00:00 ... 2009-12-15 12:00:00
Attributes:
units: $^\circ$Cdss['time']
<xarray.DataArray 'time' (time: 240)>
array([cftime.DatetimeNoLeap(1990, 1, 15, 12, 0, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(1990, 2, 14, 0, 0, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(1990, 3, 15, 12, 0, 0, 0, has_year_zero=True),
...,
cftime.DatetimeNoLeap(2009, 10, 15, 12, 0, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(2009, 11, 15, 0, 0, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(2009, 12, 15, 12, 0, 0, 0, has_year_zero=True)],
dtype=object)
Coordinates:
* time (time) object 1990-01-15 12:00:00 ... 2009-12-15 12:00:00
Attributes:
axis: T
bounds: time_bnds
standard_name: time
title: time
type: doubleThis calendar is in cftime and noLeap. Sometimes this causes issues when plotting timeseries, so just for fun we will convert to normal calendar because it’s anyway monthly.
dss['time'] = dss['time'].to_dataframe().index.to_datetimeindex()
/srv/conda/envs/notebook/lib/python3.9/site-packages/xarray/coding/times.py:360: FutureWarning: Index.ravel returning ndarray is deprecated; in a future version this will return a view on self.
sample = dates.ravel()[0]
/tmp/ipykernel_6635/4195133999.py:1: RuntimeWarning: Converting a CFTimeIndex with dates from a non-standard calendar, 'noleap', to a pandas.DatetimeIndex, which uses dates from the standard calendar. This may lead to subtle errors in operations that depend on the length of time between dates.
dss['time'] = dss['time'].to_dataframe().index.to_datetimeindex()
dss['time']
<xarray.DataArray 'time' (time: 240)>
array(['1990-01-15T12:00:00.000000000', '1990-02-14T00:00:00.000000000',
'1990-03-15T12:00:00.000000000', ..., '2009-10-15T12:00:00.000000000',
'2009-11-15T00:00:00.000000000', '2009-12-15T12:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 1990-01-15T12:00:00 ... 2009-12-15T12:00:003.3 Convert longitude:#
this data comes in 0–360 degrees, but often -180 to 180 is more convenient. So we can convert:
NOTE: Maybe you want to put this in a module? Or a package..
dss
<xarray.Dataset>
Dimensions: (member_id: 1, lat: 192, lon: 288, nbnd: 2, time: 240, lev: 32)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) datetime64[ns] 1990-01-15T12:00:00 ... 2009-12-15T12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(240, 2), meta=np.ndarray>
a_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
b_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* lev (lev) float64 -3.643 -7.595 -14.36 ... -957.5 -976.3 -992.6
lev_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
areacella (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
hurs (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 120, 192, 288), meta=np.ndarray>
tas (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 120, 192, 288), meta=np.ndarray>
a (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
b (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
mmrso4 (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 10, 32, 192, 288), meta=np.ndarray>
p0 float32 ...
ps (time, lat, lon) float32 dask.array<chunksize=(10, 192, 288), meta=np.ndarray>
T_C (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 120, 192, 288), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gnfrom util import convert360_180
(migth want to move this to a module!)
dss = convert360_180(dss)
dss['lon'].attrs['units'] = '$^\circ$ East'
dss['lon']
<xarray.DataArray 'lon' (lon: 288)>
array([-180. , -178.75, -177.5 , ..., 176.25, 177.5 , 178.75])
Coordinates:
* lon (lon) float64 -180.0 -178.8 -177.5 -176.2 ... 176.2 177.5 178.8
Attributes:
axis: X
bounds: lon_bnds
standard_name: longitude
title: Longitude
type: double
units: $^\circ$ East
valid_max: 360.0
valid_min: 0.0Notice how the labels use both the attribute “standard_name” and “units” from the dataset.
4. The easiest interpolation: select with ‘nearest’ neighboor#
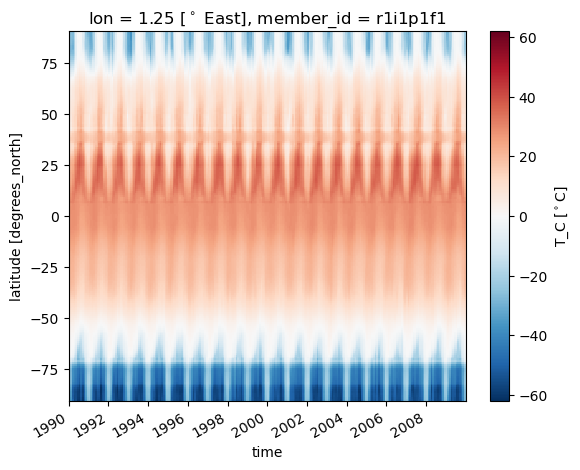
dss['T_C'].sel(lon=1.3,method='nearest').plot(x='time')
<matplotlib.collections.QuadMesh at 0x7f9572a06e20>
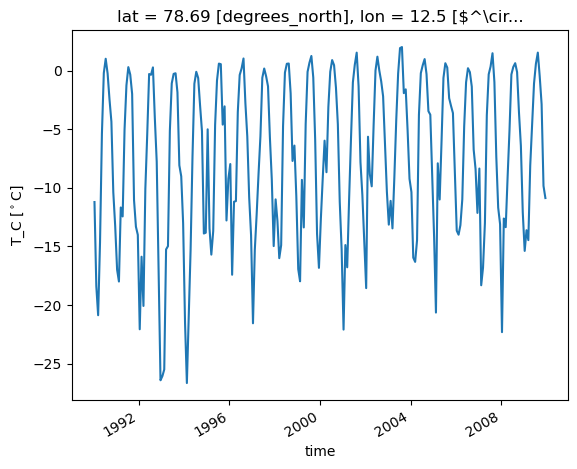
Example: let’s select zeppelin station: 78.906661, 11.889203
lat_zep =78.906661
lon_zep = 11.889203
dss['T_C'].sel(lon=lon_zep, lat=lat_zep, method='nearest').plot()
[<matplotlib.lines.Line2D at 0x7f9573b932b0>]
Super quick averaging etc#
da_so4 = dss['mmrso4']
Mean:
da_so4.mean(['time','lon'], keep_attrs=True).plot(ylim=[1000,100], yscale='log')
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Input In [34], in <cell line: 1>()
----> 1 da_so4.mean(['time','lon'], keep_attrs=True).plot(ylim=[1000,100], yscale='log')
File /srv/conda/envs/notebook/lib/python3.9/site-packages/xarray/plot/plot.py:868, in _PlotMethods.__call__(self, **kwargs)
867 def __call__(self, **kwargs):
--> 868 return plot(self._da, **kwargs)
File /srv/conda/envs/notebook/lib/python3.9/site-packages/xarray/plot/plot.py:334, in plot(darray, row, col, col_wrap, ax, hue, rtol, subplot_kws, **kwargs)
330 plotfunc = hist
332 kwargs["ax"] = ax
--> 334 return plotfunc(darray, **kwargs)
File /srv/conda/envs/notebook/lib/python3.9/site-packages/xarray/plot/plot.py:1212, in _plot2d.<locals>.newplotfunc(darray, x, y, figsize, size, aspect, ax, row, col, col_wrap, xincrease, yincrease, add_colorbar, add_labels, vmin, vmax, cmap, center, robust, extend, levels, infer_intervals, colors, subplot_kws, cbar_ax, cbar_kwargs, xscale, yscale, xticks, yticks, xlim, ylim, norm, **kwargs)
1208 raise ValueError("plt.imshow's `aspect` kwarg is not available in xarray")
1210 ax = get_axis(figsize, size, aspect, ax, **subplot_kws)
-> 1212 primitive = plotfunc(
1213 xplt,
1214 yplt,
1215 zval,
1216 ax=ax,
1217 cmap=cmap_params["cmap"],
1218 vmin=cmap_params["vmin"],
1219 vmax=cmap_params["vmax"],
1220 norm=cmap_params["norm"],
1221 **kwargs,
1222 )
1224 # Label the plot with metadata
1225 if add_labels:
File /srv/conda/envs/notebook/lib/python3.9/site-packages/xarray/plot/plot.py:1472, in pcolormesh(x, y, z, ax, xscale, yscale, infer_intervals, **kwargs)
1466 if (
1467 infer_intervals
1468 and not np.issubdtype(y.dtype, str)
1469 and (np.shape(y)[0] == np.shape(z)[0])
1470 ):
1471 if len(y.shape) == 1:
-> 1472 y = _infer_interval_breaks(y, check_monotonic=True, scale=yscale)
1473 else:
1474 # we have to infer the intervals on both axes
1475 y = _infer_interval_breaks(y, axis=1, scale=yscale)
File /srv/conda/envs/notebook/lib/python3.9/site-packages/xarray/plot/utils.py:816, in _infer_interval_breaks(coord, axis, scale, check_monotonic)
814 if scale == "log":
815 if (coord <= 0).any():
--> 816 raise ValueError(
817 "Found negative or zero value in coordinates. "
818 + "Coordinates must be positive on logscale plots."
819 )
820 coord = np.log10(coord)
822 deltas = 0.5 * np.diff(coord, axis=axis)
ValueError: Found negative or zero value in coordinates. Coordinates must be positive on logscale plots.
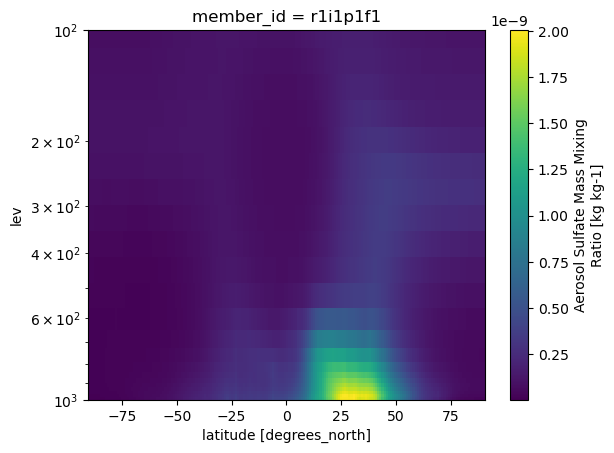
da_so4['lev'] = np.abs(da_so4['lev'].values)
da_so4.mean(['time','lon'], keep_attrs=True).plot(ylim=[1000,100], yscale='log')
<matplotlib.collections.QuadMesh at 0x7f956bdf6070>
Standard deviation
dss['T_C'].std(['time']).plot()
<matplotlib.collections.QuadMesh at 0x7f956bddaa60>
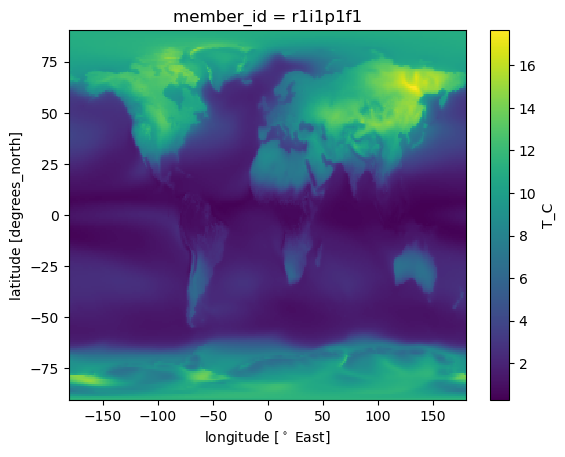
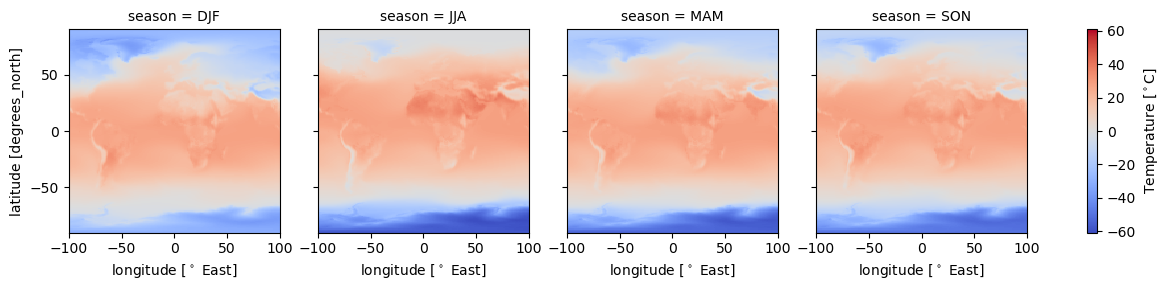
Temperature change much stronger over land than ocean…
Mask data and groupby: pick out seasons#
month = ds['time.month']
month
<xarray.DataArray 'month' (time: 1980)> array([ 1, 2, 3, ..., 10, 11, 12]) Coordinates: * time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
dss_JA = dss.where(month.isin([7,8])).mean('time')
dss_JA
<xarray.Dataset>
Dimensions: (member_id: 1, lat: 192, lon: 288, lev: 32, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 -180.0 -178.8 -177.5 -176.2 ... 176.2 177.5 178.8
* member_id (member_id) <U8 'r1i1p1f1'
* lev (lev) float64 -3.643 -7.595 -14.36 ... -957.5 -976.3 -992.6
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
a_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
b_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
lev_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
areacella (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
hurs (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
tas (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
a (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
b (lev) float64 dask.array<chunksize=(32,), meta=np.ndarray>
mmrso4 (member_id, lev, lat, lon) float32 dask.array<chunksize=(1, 32, 192, 288), meta=np.ndarray>
p0 float64 nan
ps (lat, lon) float32 dask.array<chunksize=(192, 288), meta=np.ndarray>
T_C (member_id, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>dss_season = dss.groupby('time.season').mean(keep_attrs=True)
dss_season
<xarray.Dataset>
Dimensions: (member_id: 1, season: 4, lat: 192, lon: 288, lev: 32, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 -180.0 -178.8 -177.5 -176.2 ... 176.2 177.5 178.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
b_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* lev (lev) float64 -3.643 -7.595 -14.36 ... -957.5 -976.3 -992.6
lev_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
a_bnds (lev, nbnd) float64 dask.array<chunksize=(32, 2), meta=np.ndarray>
* season (season) object 'DJF' 'JJA' 'MAM' 'SON'
Dimensions without coordinates: nbnd
Data variables:
hurs (member_id, season, lat, lon) float32 dask.array<chunksize=(1, 1, 192, 288), meta=np.ndarray>
tas (member_id, season, lat, lon) float32 dask.array<chunksize=(1, 1, 192, 288), meta=np.ndarray>
mmrso4 (member_id, season, lev, lat, lon) float32 dask.array<chunksize=(1, 1, 32, 192, 288), meta=np.ndarray>
ps (season, lat, lon) float32 dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
T_C (member_id, season, lat, lon) float32 dask.array<chunksize=(1, 1, 192, 288), meta=np.ndarray>
areacella (season, member_id, lat, lon) float32 dask.array<chunksize=(4, 1, 192, 288), meta=np.ndarray>
a (season, lev) float64 dask.array<chunksize=(4, 32), meta=np.ndarray>
b (season, lev) float64 dask.array<chunksize=(4, 32), meta=np.ndarray>
p0 (season) float32 1e+05 1e+05 1e+05 1e+05
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gndss_season['T_C'].plot(col='season')
<xarray.plot.facetgrid.FacetGrid at 0x7f95640ef190>
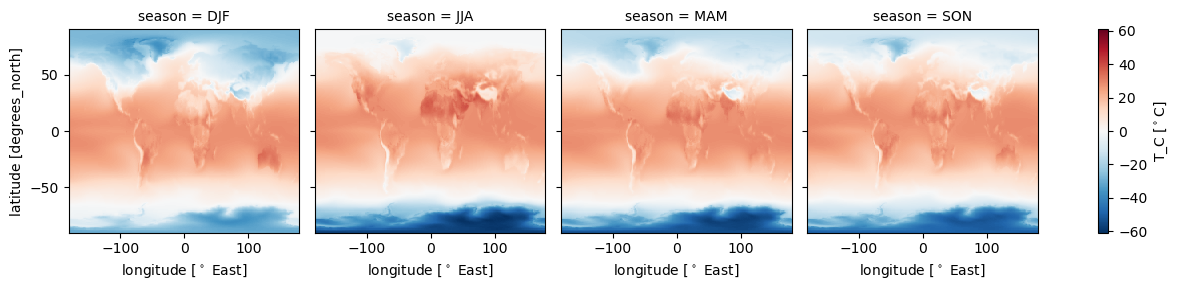
Controle the plot visuals:#
dss_season['T_C'].plot(col='season',
cmap = 'coolwarm',#ax=ax,
xlim=[-100,100],
cbar_kwargs={'label':'Temperature [$^\circ$C]'})
<xarray.plot.facetgrid.FacetGrid at 0x7f95644a3430>
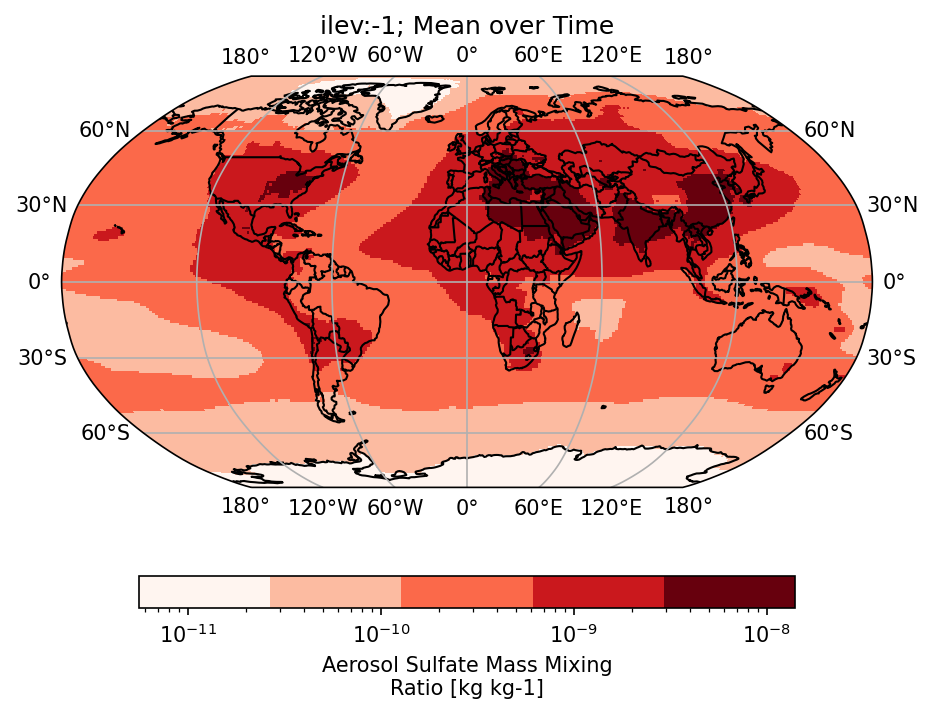
5. Plotting with cartopy#
import cartopy as cy
import cartopy.crs as ccrs
da_plt = dss['mmrso4'].isel(lev=-1).mean('time', keep_attrs=True).squeeze()#('member_id')
da_plt
<xarray.DataArray 'mmrso4' (lat: 192, lon: 288)>
dask.array<getitem, shape=(192, 288), dtype=float32, chunksize=(192, 288), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 -180.0 -178.8 -177.5 -176.2 ... 176.2 177.5 178.8
member_id <U8 'r1i1p1f1'
lev float64 -992.6
Attributes:
cell_measures: area: areacella
cell_methods: area: time: mean
comment: Dry mass of sulfate (SO4) in aerosol particles as a fract...
description: Dry mass of sulfate (SO4) in aerosol particles as a fract...
frequency: mon
id: mmrso4
long_name: Aerosol Sulfate Mass Mixing Ratio
mipTable: AERmon
out_name: mmrso4
prov: AERmon ((isd.003))
realm: aerosol
standard_name: mass_fraction_of_sulfate_dry_aerosol_particles_in_air
time: time
time_label: time-mean
time_title: Temporal mean
title: Aerosol Sulfate Mass Mixing Ratio
type: real
units: kg kg-1
variable_id: mmrso4from matplotlib.colors import LogNorm
f,ax = plt.subplots(1,1,dpi=150,subplot_kw={'projection':ccrs.Robinson()})
da_plt.plot.pcolormesh(
cmap = plt.get_cmap('Reds'),
ax=ax,
norm = LogNorm(),
cbar_kwargs={
#'label':'Wind Speed [m/s]',
'orientation':'horizontal',
'shrink':.8
},
transform=ccrs.PlateCarree(),
#x='lon',y='lat',
levels = 6
)
ax.set_title('ilev:-1; Mean over Time')
ax.coastlines()
gl = ax.gridlines(draw_labels=True)
#gl.xlabels_top = False
#gl.ylabels_right = False
#gl.top_labels(False)
ax.add_feature(cy.feature.BORDERS);
f.tight_layout()
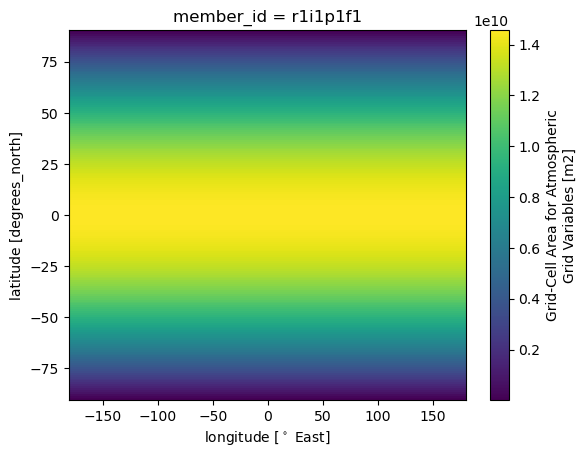
6. Global average: weighted means!#
dss['T_C'].mean().compute()
<xarray.DataArray 'T_C' ()> array(5.447856, dtype=float32)
dss['T_C'].weighted(dss['areacella']).mean().compute()
<xarray.DataArray 'T_C' ()> array(14.715081, dtype=float32)
dss['areacella'].plot()
<matplotlib.collections.QuadMesh at 0x7f95643c1670>
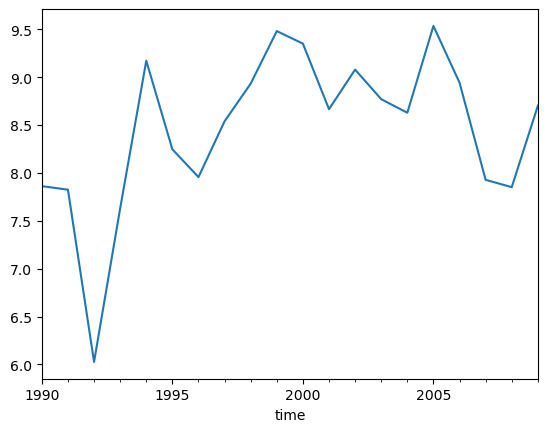
7. Convert to pandas & do some random fun stuff:#
Maybe we e.g. want to compare with a station, or just use some of the considerable functionalities available from pandas. It’s easy to convert back and forth between xarray and pandas:
A lot of these functions also exist in xarray!Pick out station:#
lat_tjarno = 58.9
lon_tjarno = 11.1
# pick out surface
ds_surf =dss.isel(lev=-1)
ds_tjarno = ds_surf.sel(lat=lat_tjarno, lon = lon_tjarno, method ='nearest')
ds_tjarno
<xarray.Dataset>
Dimensions: (member_id: 1, nbnd: 2, time: 240)
Coordinates:
lat float64 58.9
lon float64 11.25
lon_bnds (nbnd) float64 dask.array<chunksize=(2,), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) datetime64[ns] 1990-01-15T12:00:00 ... 2009-12-15T12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(240, 2), meta=np.ndarray>
a_bnds (nbnd) float64 dask.array<chunksize=(2,), meta=np.ndarray>
b_bnds (nbnd) float64 dask.array<chunksize=(2,), meta=np.ndarray>
lev float64 -992.6
lev_bnds (nbnd) float64 dask.array<chunksize=(2,), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
areacella (member_id) float32 dask.array<chunksize=(1,), meta=np.ndarray>
hurs (member_id, time) float32 dask.array<chunksize=(1, 120), meta=np.ndarray>
tas (member_id, time) float32 dask.array<chunksize=(1, 120), meta=np.ndarray>
a float64 dask.array<chunksize=(), meta=np.ndarray>
b float64 dask.array<chunksize=(), meta=np.ndarray>
mmrso4 (member_id, time) float32 dask.array<chunksize=(1, 10), meta=np.ndarray>
p0 float32 ...
ps (time) float32 dask.array<chunksize=(10,), meta=np.ndarray>
T_C (member_id, time) float32 dask.array<chunksize=(1, 120), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gnvl = ['mmrso4','hurs','tas','T_C']
ds_tjarno[vl]
<xarray.Dataset>
Dimensions: (member_id: 1, time: 240)
Coordinates:
lat float64 58.9
lon float64 11.25
* member_id (member_id) <U8 'r1i1p1f1'
* time (time) datetime64[ns] 1990-01-15T12:00:00 ... 2009-12-15T12:00:00
lev float64 -992.6
Data variables:
mmrso4 (member_id, time) float32 dask.array<chunksize=(1, 10), meta=np.ndarray>
hurs (member_id, time) float32 dask.array<chunksize=(1, 120), meta=np.ndarray>
tas (member_id, time) float32 dask.array<chunksize=(1, 120), meta=np.ndarray>
T_C (member_id, time) float32 dask.array<chunksize=(1, 120), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/75033260-7bd2-46b9-9d98-3456f176d588
version_id: v20190308
intake_esm_varname: ['areacella']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.fx.gnResample:#
df_tjarno = ds_tjarno[vl].squeeze().to_dataframe()
df_tjarno
| mmrso4 | hurs | tas | T_C | lat | lon | member_id | lev | |
|---|---|---|---|---|---|---|---|---|
| time | ||||||||
| 1990-01-15 12:00:00 | 9.264967e-10 | 89.233849 | 275.603851 | 2.453857 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
| 1990-02-14 00:00:00 | 7.049255e-10 | 82.765656 | 271.453064 | -1.696930 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
| 1990-03-15 12:00:00 | 1.147899e-09 | 83.034279 | 276.359680 | 3.209686 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
| 1990-04-15 00:00:00 | 6.494133e-10 | 73.870636 | 279.862518 | 6.712524 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
| 1990-05-15 12:00:00 | 2.289224e-09 | 74.073692 | 283.468597 | 10.318604 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2009-08-15 12:00:00 | 5.304504e-10 | 77.305382 | 290.360901 | 17.210907 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
| 2009-09-15 00:00:00 | 5.245591e-10 | 75.595207 | 287.631470 | 14.481476 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
| 2009-10-15 12:00:00 | 4.940789e-10 | 83.765961 | 283.187012 | 10.037018 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
| 2009-11-15 00:00:00 | 1.800878e-10 | 88.817947 | 276.953217 | 3.803223 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
| 2009-12-15 12:00:00 | 3.002016e-10 | 91.949974 | 275.946564 | 2.796570 | 58.900524 | 11.25 | r1i1p1f1 | -992.556095 |
240 rows × 8 columns
df_yearly = df_tjarno.resample('Y').mean()#.plot()
df_yearly['T_C'].plot()
<AxesSubplot:xlabel='time'>
df_tjarno['season'] = df_tjarno.to_xarray()['time.season']
for s in ['MAM','JJA','SON','DJF']:
df_tjarno[df_tjarno['season']==s]['T_C'].plot.hist(alpha=0.5, bins=20, label=s)
plt.legend()
<matplotlib.legend.Legend at 0x7f953c140d90>
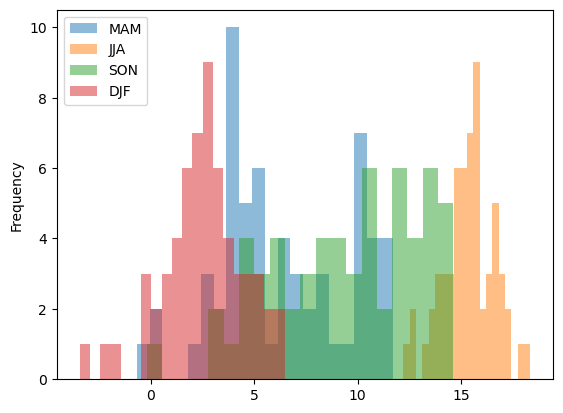
df = dss.isel(lev=-1)[vl].to_dataframe()
df_ri = df.reset_index()
df_ri.head()
| member_id | time | lat | lon | mmrso4 | hurs | tas | T_C | lev | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | r1i1p1f1 | 1990-01-15 12:00:00 | -90.0 | -180.00 | 2.049096e-11 | 98.689842 | 242.897797 | -30.252197 | -992.556095 |
| 1 | r1i1p1f1 | 1990-01-15 12:00:00 | -90.0 | -178.75 | 2.049095e-11 | 98.675537 | 242.903198 | -30.246796 | -992.556095 |
| 2 | r1i1p1f1 | 1990-01-15 12:00:00 | -90.0 | -177.50 | 2.049095e-11 | 98.675537 | 242.903198 | -30.246796 | -992.556095 |
| 3 | r1i1p1f1 | 1990-01-15 12:00:00 | -90.0 | -176.25 | 2.049095e-11 | 98.675537 | 242.903198 | -30.246796 | -992.556095 |
| 4 | r1i1p1f1 | 1990-01-15 12:00:00 | -90.0 | -175.00 | 2.049095e-11 | 98.675537 | 242.903198 | -30.246796 | -992.556095 |
lets do something unnecesarily complicated :D#
qcut, cut#
qcut splits the data into quantile ranges
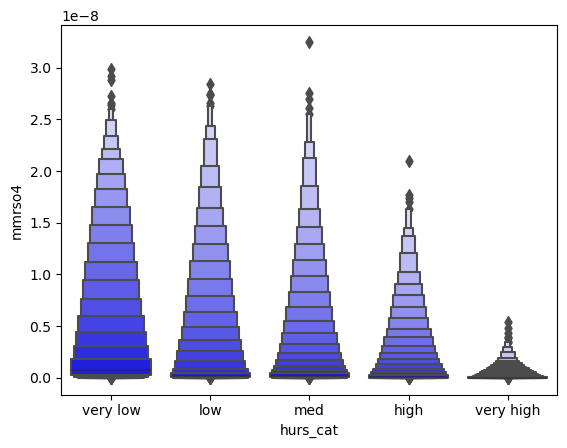
df_ri['hurs_cat'] = pd.qcut(df_ri['hurs'],
q=[0.05,0.17, 0.34,0.66, 0.83,0.95],
labels=['very low','low','med','high','very high'])
Cut cuts into categories
df_ri['lat_cat'] = pd.cut(df_ri['lat'], [-90,-60,-30,0,30,60,90],
labels=['S polar','S mid','S tropics', 'N tropic', 'N mid','N polar'])
df_ri.groupby('lat_cat').mean()
| lat | lon | mmrso4 | hurs | tas | T_C | lev | |
|---|---|---|---|---|---|---|---|
| lat_cat | |||||||
| S polar | -74.921466 | -0.625 | 2.556384e-11 | 95.399734 | 247.324890 | -25.825100 | -992.556095 |
| S mid | -45.235602 | -0.625 | 1.597070e-10 | 79.675735 | 282.781799 | 9.631801 | -992.556095 |
| S tropics | -15.078534 | -0.625 | 3.580959e-10 | 76.560928 | 297.261230 | 24.111250 | -992.556095 |
| N tropic | 15.078534 | -0.625 | 1.236901e-09 | 73.094543 | 298.772369 | 25.622366 | -992.556095 |
| N mid | 45.235602 | -0.625 | 1.287050e-09 | 73.039864 | 282.893677 | 9.743673 | -992.556095 |
| N polar | 75.392670 | -0.625 | 1.318637e-10 | 95.613152 | 263.219543 | -9.930449 | -992.556095 |
sns.boxenplot(x="lat_cat", y="hurs",
color="b",
scale="linear", data=df_ri)
<AxesSubplot:xlabel='lat_cat', ylabel='hurs'>
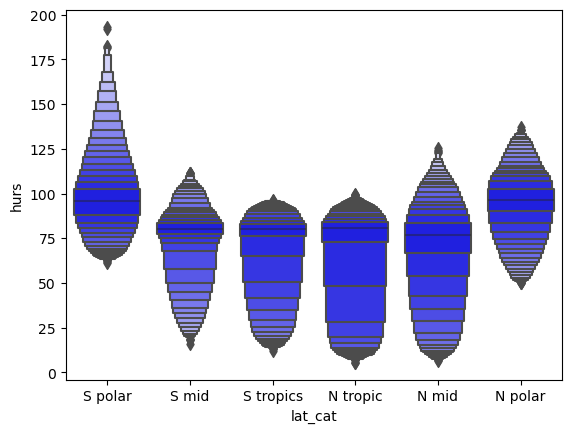
sns.boxenplot(x="hurs_cat", y="mmrso4",
color="b",
scale="linear", data=df_ri,
)
<AxesSubplot:xlabel='hurs_cat', ylabel='mmrso4'>
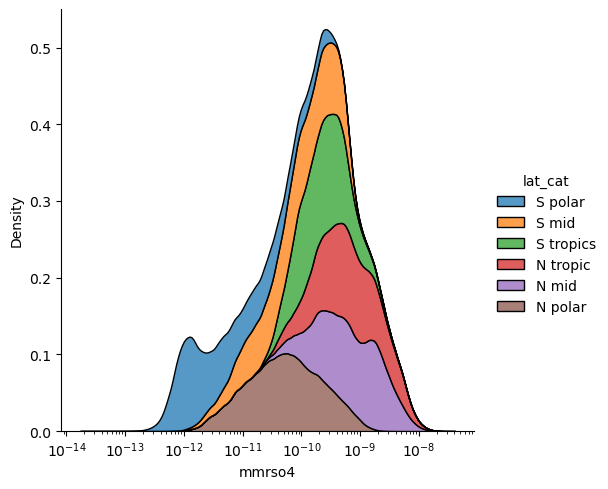
sns.displot(x="mmrso4",hue = 'lat_cat',log_scale=True,kind='kde',
data=df_ri, multiple="stack")
<seaborn.axisgrid.FacetGrid at 0x7f953c101fa0>
Convert back to xarray if we need:#
ds_new = df_ri.set_index(['time','lat','lon']).to_xarray()
ds_new
<xarray.Dataset>
Dimensions: (time: 240, lat: 192, lon: 288)
Coordinates:
* time (time) datetime64[ns] 1990-01-15T12:00:00 ... 2009-12-15T12:00:00
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 -180.0 -178.8 -177.5 -176.2 ... 176.2 177.5 178.8
Data variables:
member_id (time, lat, lon) object 'r1i1p1f1' 'r1i1p1f1' ... 'r1i1p1f1'
mmrso4 (time, lat, lon) float32 2.049e-11 2.049e-11 ... 5.151e-11
hurs (time, lat, lon) float32 98.69 98.68 98.68 ... 106.9 106.9 106.9
tas (time, lat, lon) float32 242.9 242.9 242.9 ... 250.9 250.9 250.9
T_C (time, lat, lon) float32 -30.25 -30.25 -30.25 ... -22.28 -22.28
lev (time, lat, lon) float64 -992.6 -992.6 -992.6 ... -992.6 -992.6
hurs_cat (time, lat, lon) object 'very high' 'very high' ... nan nan
lat_cat (time, lat, lon) object nan nan nan ... 'N polar' 'N polar'mask by category#
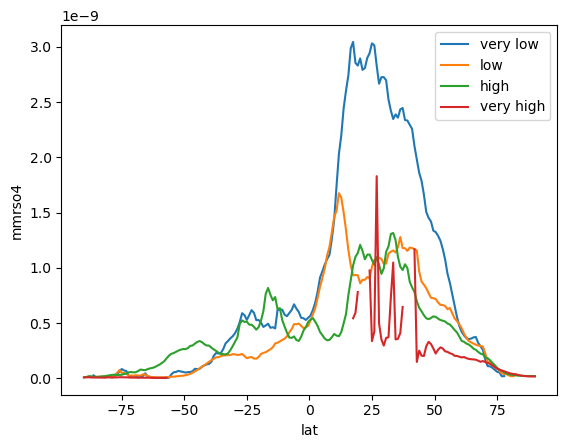
ds_new.where(ds_new['hurs_cat']=='very low').mean(['time','lon'])['mmrso4'].plot(label='very low')#vmin = 0, vmax = 1.5e-8)
ds_new.where(ds_new['hurs_cat']=='low').mean(['time','lon'])['mmrso4'].plot(label='low')#vmin = 0, vmax = 1.5e-8)
ds_new.where(ds_new['hurs_cat']=='high').mean(['time','lon'])['mmrso4'].plot(label='high')#vmin = 0, vmax = 1.5e-8)
ds_new.where(ds_new['hurs_cat']=='very high').mean(['time','lon'], keep_attrs=True)['mmrso4'].plot(label='very high')#vmin = 0, vmax = 1.5e-8)
plt.legend()
<matplotlib.legend.Legend at 0x7f9508f85b20>