Interactive plotting with HoloViews
Contents
Interactive plotting with HoloViews#
Context#
We will be using HoloViews, a tool part of the HoloViz ecosystem, with Xarray to visualize the near-surface temperature from CMIP6 CESM2 (monthly dataset).
Data#
We will visualize the same data that have been used in the previous episode.
If the dataset is not present in the same folder as this Jupyter notebook, it will be downloaded from zenodo using pooch, a very handy python-based library to download and cache your data files locally (see further info here).
import pooch
tas_file = pooch.retrieve(
url="https://zenodo.org/record/7181714/files/CMIP6_NCAR_CESM2_historical_amon_gn.nc",
known_hash="md5:5f86251e5bc5ef9b86a3a86cd06a536b",
path=f".",
)
Setup#
This episode uses the following main Python packages:
pooch [USR+20]
rioxarray [SBR+22]
matplotlib [Hun07]
cartopy [MetOffice15]
hvplot [RSB+20]
geopandas [JdBF+20]
Please install these packages if not already available in your Python environment.
Packages#
In this episode, Python packages are imported when we start to use them. However, for best software practices, we recommend you to install and import all the necessary libraries at the top of your Jupyter notebook.
import xarray as xr
Open local dataset#
tas_ds = xr.open_dataset(tas_file)
Tip
If you get an error with the previous command, check the previous episode where the input file some_hash-CMIP6_NCAR_CESM2_historical_amon_gn.nc is downloaded locally and it is in the same directory as your Jupyter Notebook.
tas_ds
<xarray.Dataset>
Dimensions: (lat: 192, nbnd: 2, lon: 288, member_id: 1, time: 1980)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
lat_bnds (lat, nbnd) float32 -90.0 -89.53 -89.53 ... 89.53 89.53 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float32 -0.625 0.625 0.625 ... 358.1 358.1 359.4
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
time_bnds (time, nbnd) object 1850-01-01 00:00:00 ... 2015-01-01 00:00:00
* member_id (member_id) object 'r1i1p1f1'
Dimensions without coordinates: nbnd
Data variables:
tas (member_id, time, lat, lon) float32 ...
Attributes: (12/50)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/d9a7225a-49c3-4470-b7ab-a8180926f839
version_id: v20190308
intake_esm_varname: tas
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.Amon.gnClipping data according to a polygon#
One of the basic concepts in GIS is to clip data using a vector geometry. Xarray is not directly capable of dealing with vectors but thanks to Rioxarray that can be easily achieved. Rioxarray extends Xarray with most of the features that Rasterio (GDAL) brings.
Read a shapefile with the Area Of Interest (AOI)#
import geopandas as gpd
We define the area of interest using the Global Administrative Unit Layers GAUL G2015_2014 provided by FAO-UN (see Documentation).
GeoPandas, a python-based library extending the capabilities of Pandas to deal with geometry and spatial operations, will help to manage geodata.
The official data distribution from FAO is through the WFS service (see below how to retrieve data):
GAUL = gpd.read_file('https://data.apps.fao.org/map/gsrv/gsrv1/gaul/wfs?'
'service=WFS&version=2.0.0&'
'Request=GetFeature&'
'TypeNames=gaul:g2015_2014_2&'
'srsName=EPSG%3A4326&'
'maxFeatures=2500&'
'outputFormat=json')
Unfortunately it seems that the service is pretty slow. As an alternative to this approach the JRC MARS unit is distributing the original dataset that was in shapefile format. To accelerate the fetch we highly recommend to follow this approach.
For the training course, we also created a tiny file containing information about Norway only.
try:
GAUL = gpd.read_file('Norway.geojson')
except:
GAUL = gpd.read_file('zip+https://mars.jrc.ec.europa.eu/asap/files/gaul1_asap.zip')
Data are organized in a tabular structure. For each element an index, data (made of columns) and a geometry are defined.
Geometries are defined through shapely geometry objects with three different basic classes:
Points and Multi-Points
Lines and Multi-Lines
Polygons and Multi-Polygons
GAUL.head(5)
| asap1_id | name1 | name1_shr | name0 | asap0_id | name0_shr | km2_tot | km2_crop | km2_range | an_crop | an_range | water_lim | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1238 | Hordaland | Hordaland | Norway | 173 | Norway | 14021 | 631 | 81 | 1 | 0 | 0 | MULTIPOLYGON (((5.67021 60.69386, 5.67357 60.6... |
| 1 | 1446 | Nord-troendelag | Nord-troendelag | Norway | 173 | Norway | 22315 | 1344 | 9 | 1 | 0 | 0 | MULTIPOLYGON (((11.93731 65.10981, 11.88333 65... |
| 2 | 1472 | Soer-Troendelag | Soer-Troendelag | Norway | 173 | Norway | 18441 | 1313 | 4 | 1 | 0 | 0 | MULTIPOLYGON (((9.74026 63.62652, 9.72871 63.6... |
| 3 | 1426 | Oestfold | Oestfold | Norway | 173 | Norway | 4128 | 1030 | 0 | 1 | 0 | 0 | POLYGON ((11.94171 59.69688, 11.89933 59.69182... |
| 4 | 1496 | Vestfold | Vestfold | Norway | 173 | Norway | 2232 | 623 | 2 | 1 | 0 | 0 | POLYGON ((10.39417 59.66410, 10.42124 59.62203... |
In the cell below, we subset the polygon geometry in which the name0 field equals to Norway.
AOI_name = 'Norway'
AOI = GAUL[GAUL.name0 == AOI_name]
AOI_poly = AOI.geometry
AOI_poly
0 MULTIPOLYGON (((5.67021 60.69386, 5.67357 60.6...
1 MULTIPOLYGON (((11.93731 65.10981, 11.88333 65...
2 MULTIPOLYGON (((9.74026 63.62652, 9.72871 63.6...
3 POLYGON ((11.94171 59.69688, 11.89933 59.69182...
4 POLYGON ((10.39417 59.66410, 10.42124 59.62203...
5 MULTIPOLYGON (((22.69320 70.36594, 22.70137 70...
6 POLYGON ((8.13729 60.19619, 8.16250 60.19416, ...
7 MULTIPOLYGON (((13.97719 68.35144, 13.99107 68...
8 MULTIPOLYGON (((8.01763 63.32659, 7.97557 63.3...
9 POLYGON ((5.23273 62.19275, 5.25814 62.17775, ...
10 MULTIPOLYGON (((15.76350 68.74171, 15.77052 68...
11 POLYGON ((8.29320 61.08945, 8.31227 61.07882, ...
12 MULTIPOLYGON (((10.64400 59.90339, 10.63911 59...
13 POLYGON ((8.87194 62.37852, 8.89526 62.35425, ...
14 POLYGON ((11.13333 62.65833, 11.14690 62.64664...
15 POLYGON ((7.26296 59.65463, 7.33571 59.63810, ...
16 POLYGON ((7.02216 59.18047, 7.09886 59.17007, ...
17 POLYGON ((10.91103 59.92944, 10.89463 59.90425...
18 POLYGON ((6.51667 59.84167, 6.58167 59.83139, ...
Name: geometry, dtype: geometry
In a second step we set a EPSG:4326 Geodetic coordinate reference system to the polygon geometry. To achieve this we need to rely on rioxarray that extends xarray with the rasterio capabilities. The rio accessor is activated through importing rioxarray as has been done at the top.
tas_ds.rio.write_crs(4326, inplace=True)
<xarray.Dataset>
Dimensions: (lat: 192, nbnd: 2, lon: 288, member_id: 1, time: 1980)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
lat_bnds (lat, nbnd) float32 -90.0 -89.53 -89.53 ... 89.53 89.53 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float32 -0.625 0.625 0.625 ... 358.1 358.1 359.4
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
time_bnds (time, nbnd) object 1850-01-01 00:00:00 ... 2015-01-01 00:00:00
* member_id (member_id) object 'r1i1p1f1'
spatial_ref int64 0
Dimensions without coordinates: nbnd
Data variables:
tas (member_id, time, lat, lon) float32 ...
Attributes: (12/50)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
... ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/d9a7225a-49c3-4470-b7ab-a8180926f839
version_id: v20190308
intake_esm_varname: tas
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.Amon.gnOnce this has been done we can clip the data with the polygon that has been obtained through geopandas at the beginning of the notebook.
tas_AOI = tas_ds.tas.rio.clip(AOI_poly, crs=4326)
tas_AOI
<xarray.DataArray 'tas' (member_id: 1, time: 1980, lat: 13, lon: 21)>
array([[[[ nan, 268.18625, 263.21762, ..., nan, nan,
nan],
[ nan, 261.4894 , 259.58054, ..., nan, nan,
nan],
[270.17227, 260.38303, 257.93915, ..., nan, nan,
nan],
...,
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., 262.4566 , 262.70773,
262.97894]],
[[ nan, 274.94598, 272.1838 , ..., nan, nan,
nan],
[ nan, 272.17087, 270.1366 , ..., nan, nan,
nan],
[276.09476, 271.16046, 269.08408, ..., nan, nan,
nan],
...
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., 268.94943, 268.97186,
269.03262]],
[[ nan, 280.14944, 277.7591 , ..., nan, nan,
nan],
[ nan, 276.78476, 274.67453, ..., nan, nan,
nan],
[280.1642 , 275.19962, 273.14044, ..., nan, nan,
nan],
...,
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., 268.04092, 268.38055,
268.77368]]]], dtype=float32)
Coordinates:
* lat (lat) float64 58.9 59.84 60.79 61.73 ... 68.32 69.27 70.21
* lon (lon) float64 5.0 6.25 7.5 8.75 10.0 ... 26.25 27.5 28.75 30.0
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
* member_id (member_id) object 'r1i1p1f1'
spatial_ref int64 0
Attributes:
cell_measures: area: areacella
cell_methods: area: time: mean
comment: near-surface (usually, 2 meter) air temperature
description: near-surface (usually, 2 meter) air temperature
frequency: mon
id: tas
long_name: Near-Surface Air Temperature
mipTable: Amon
out_name: tas
prov: Amon ((isd.003))
realm: atmos
standard_name: air_temperature
time: time
time_label: time-mean
time_title: Temporal mean
title: Near-Surface Air Temperature
type: real
units: K
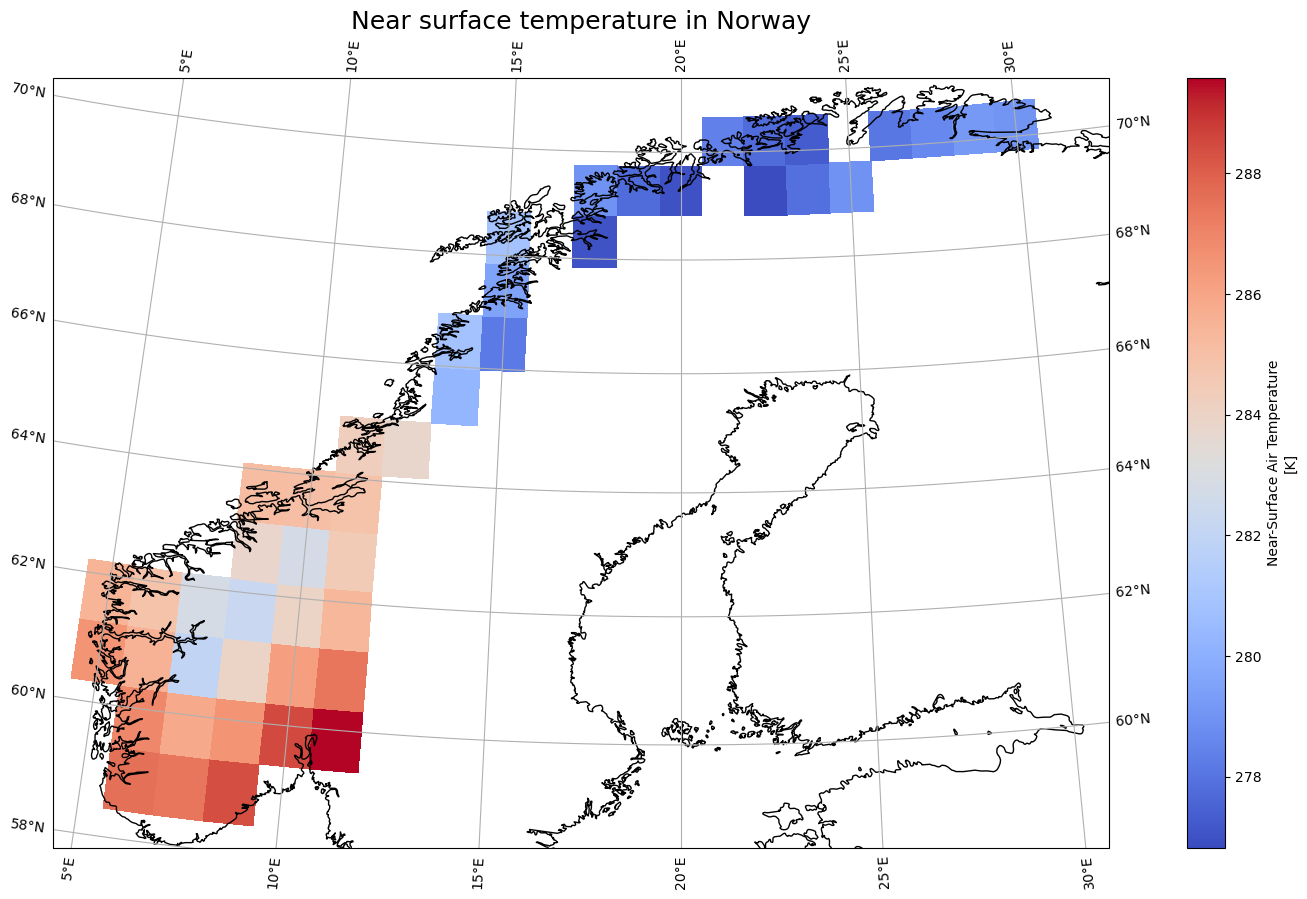
variable_id: tasVisualize with matplotlib#
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
fig = plt.figure(1, figsize=[20, 10])
# We're using cartopy and are plotting in PlateCarree projection
# (see documentation on cartopy)
ax = plt.subplot(1, 1, 1, projection=ccrs.AlbersEqualArea(central_longitude=20.0, central_latitude=40.0))
#ax.set_extent([15.5, 27.5, 36, 41], crs=ccrs.PlateCarree()) # lon1 lon2 lat1 lat2
ax.coastlines(resolution='10m')
ax.gridlines(draw_labels=True)
tas_AOI.sel(time='1980-06-15').plot(ax=ax, transform=ccrs.PlateCarree(), cmap="coolwarm")
# One way to customize your title
plt.title("Near surface temperature in Norway", fontsize=18)
Text(0.5, 1.0, 'Near surface temperature in Norway')
Visualization with HoloViews#
import holoviews as hv
import hvplot.xarray
import hvplot.pandas
Plotting data through the HoloViews back-end thanks to the hvplot that acts as high-level plotting API.
tas_AOI.isel(time=0, member_id=0).hvplot(cmap="coolwarm", width=1000, height=1000)
Having a look to data distribution can reveal a lot about the data.
tas_AOI[0].hvplot.hist(cmap="coolwarm",bins=25, width=800, height=700)
Multi-plots using groupby#
To be able to visualize interactively all the different available times, we can use groupby time.
tas_AOI.isel(member_id=0).hvplot(groupby ='time', cmap="coolwarm", width=800, height=700)
We can add a histogram to the visualization.
tas_AOI.isel(member_id=0).hvplot(groupby='time', cmap='coolwarm', width=800, height=700 ).hist()
Plot data on a particular projection#
plot = tas_AOI.isel(time=0, member_id=0).hvplot.quadmesh(x='lon', y='lat'
, rasterize=True
, geo=True
, global_extent=False
, projection=ccrs.Orthographic(30, 90)
, project=True
, cmap='coolwarm'
, coastline='50m'
, frame_width=400
, title="Near-surface Temperature over Norway (CMIP6 CESM2)"
)
WARNING:param.main: Calling the .opts method with options broken down by options group (i.e. separate plot, style and norm groups) is deprecated. Use the .options method converting to the simplified format instead or use hv.opts.apply_groups for backward compatibility.
plot
Plot a single point over the time dimension#
tas_AOI.isel(member_id=0).sel(lat=60, lon=10.75, method='nearest').hvplot(ylim=(260, 293))
WARNING:param.CurvePlot02669: Converting cftime.datetime from a non-standard calendar (noleap) to a standard calendar for plotting. This may lead to subtle errors in formatting dates, for accurate tick formatting switch to the matplotlib backend.
- Rioxarray for clipping over an area of interest
- Interactive plot with HoloViews and Xarray
References#
- Kog95
F.N. Kogan. Application of vegetation index and brightness temperature for drought detection. Advances in Space Research, 15(11):91–100, 1995. Natural Hazards: Monitoring and Assessment Using Remote Sensing Technique. URL: https://www.sciencedirect.com/science/article/pii/027311779500079T, doi:https://doi.org/10.1016/0273-1177(95)00079-T.
Packages citation#
- HH17
S. Hoyer and J. Hamman. Xarray: N-D labeled arrays and datasets in Python. Journal of Open Research Software, 2017. URL: https://doi.org/10.5334/jors.148, doi:10.5334/jors.148.
- Hun07
J. D. Hunter. Matplotlib: a 2d graphics environment. Computing in Science & Engineering, 9(3):90–95, 2007. doi:10.1109/MCSE.2007.55.
- JdBF+20
Kelsey Jordahl, Joris Van den Bossche, Martin Fleischmann, Jacob Wasserman, James McBride, Jeffrey Gerard, Jeff Tratner, Matthew Perry, Adrian Garcia Badaracco, Carson Farmer, Geir Arne Hjelle, Alan D. Snow, Micah Cochran, Sean Gillies, Lucas Culbertson, Matt Bartos, Nick Eubank, maxalbert, Aleksey Bilogur, Sergio Rey, Christopher Ren, Dani Arribas-Bel, Leah Wasser, Levi John Wolf, Martin Journois, Joshua Wilson, Adam Greenhall, Chris Holdgraf, Filipe, and François Leblanc. Geopandas/geopandas: v0.8.1. July 2020. URL: https://doi.org/10.5281/zenodo.3946761, doi:10.5281/zenodo.3946761.
- RSB+20
Philipp Rudiger, Jean-Luc Stevens, James A. Bednar, Bas Nijholt, Andrew, Chris B, Achim Randelhoff, Jon Mease, Vasco Tenner, maxalbert, Markus Kaiser, ea42gh, Jordan Samuels, stonebig, Florian LB, Andrew Tolmie, Daniel Stephan, Scott Lowe, John Bampton, henriqueribeiro, Irv Lustig, Julia Signell, Justin Bois, Leopold Talirz, Lukas Barth, Maxime Liquet, Ram Rachum, Yuval Langer, arabidopsis, and kbowen. Holoviz/holoviews: version 1.13.3. June 2020. URL: https://doi.org/10.5281/zenodo.3904606, doi:10.5281/zenodo.3904606.
- SBR+22
Alan D. Snow, David Brochart, Martin Raspaud, Ray Bell, RichardScottOZ, Taher Chegini, Alessandro Amici, Rémi Braun, Andrew Annex, David Hoese, Fred Bunt, GBallesteros, Joe Hamman, Markus Zehner, Mauricio Cordeiro, Scott Henderson, Seth Miller, The Gitter Badger, Tom Augspurger, apiwat-chantawibul, and pmallas. Corteva/rioxarray: 0.11.1 release. April 2022. URL: https://doi.org/10.5281/zenodo.6478182, doi:10.5281/zenodo.6478182.
- USR+20
Leonardo Uieda, Santiago Rubén Soler, Rémi Rampin, Hugo van Kemenade, Matthew Turk, Daniel Shapero, Anderson Banihirwe, and John Leeman. Pooch: a friend to fetch your data files. Journal of Open Source Software, 5(45):1943, 2020. URL: https://doi.org/10.21105/joss.01943, doi:10.21105/joss.01943.
- MetOffice15
Met Office. Cartopy: a cartographic python library with a matplotlib interface. Exeter, Devon, 2010 - 2015. URL: http://scitools.org.uk/cartopy.


